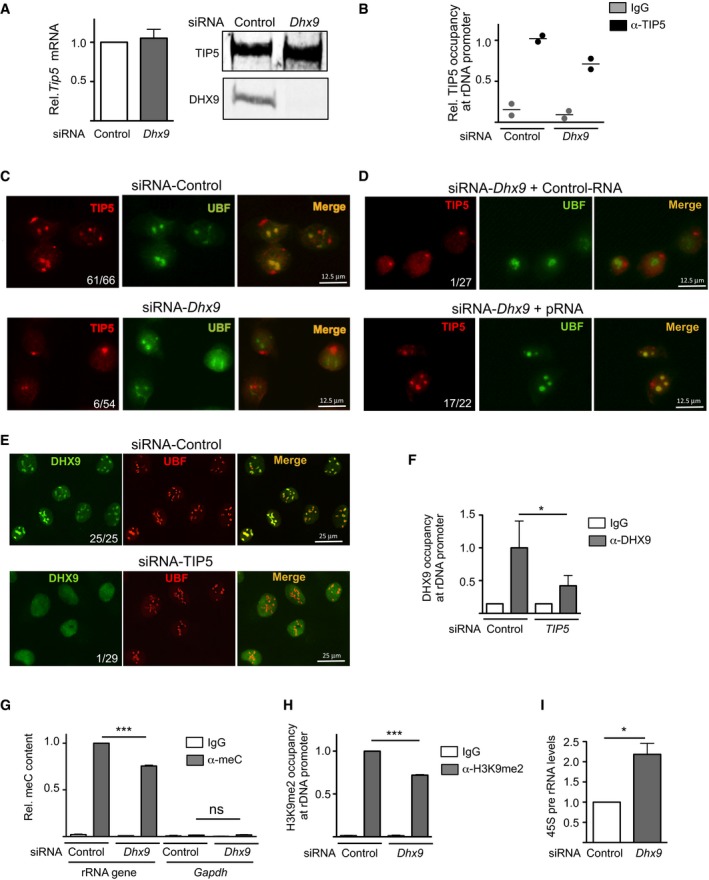
DHX9 knockdown does not affect TIP5 levels. mRNA and protein levels of TIP5 in NIH 3T3 cells depleted of DHX9 by siRNA. TIP5 mRNA values were normalized to GAPDH mRNA and to siRNA‐control cells. Values (mean ± SD) are from three independent experiments.
DHX9 is required for the association of TIP5 with rRNA genes. ChIP analysis of TIP5 occupancy at rRNA genes in NIH 3T3 cells depleted of DHX9 by siRNA. Data from two independent experiments are represented as bound over input and normalized to siRNA‐Control cells. Scatter plot represents the values and the mean of two independent experiments.
DHX9 is required for the localization of TIP5 in nucleoli. Immunofluorescence with anti‐TIP5 and anti‐UBF of NIH 3T3 cells treated with siRNA‐Control or‐DHX9. Numbers refer to cells showing TIP5 nucleolar localization relative to the number of analyzed cells.
Retention of TIP5 in nucleoli depends on DHX9‐mediated production of mature pRNA. Immunofluorescence with anti‐TIP5 and anti‐UBF of DHX9‐depleted NIH 3T3 cells transfected with Control‐RNA or pRNA.
Nucleolar localization of DHX9 depends on TIP5. Immunofluorescence with anti‐DHX9 and anti‐UBF of U2OS cells treated with siRNA‐Control or siRNA‐TIP5. Numbers refer to cells showing DHX9 nucleolar localization relative to the number of analyzed cells.
TIP5 is required for the association of DHX9 with rRNA genes. ChIP analysis of DHX9 occupancy at rRNA genes in U2OS cells depleted of TIP5 by siRNA. Values (mean ± SD) from three independent experiments are represented as bound over input and normalized to siRNA‐Control cells.
DHX9 is required for rRNA gene silencing. Methylated DNA immunoprecipitation (MeDIP) analysis of rRNA gene promoter using anti‐5mC antibodies in NIH 3T3 cells upon DHX9 knockdown. Enrichments were calculated relative to input and normalized to rRNA genes in control cells. Low enrichment of GAPDH sequences (free of CpG methylation) ensures for the specificity of the measurement. Values (mean ± SD) are from three independent experiments.
ChIP analysis of H3K9me2 at rRNA gene promoter in NIH 3T3 cells upon DHX9 knockdown. Enrichments were calculated relative to input and normalized to control cells. Values (mean ± SD) are from three independent experiments.
Knockdown of DHX9 upregulates rRNA transcription. RT–qPCR of 45S pre‐rRNA levels in NIH 3T3 cells upon DHX9 knockdown. Values (mean ± SD) from three independent experiments were normalized to GAPDH mRNA and to control cells.
< 0.001; ns, non‐significant).