Abstract
In the present study, the function of S100 calcium binding protein P (S100P) in the C666-1 nasopharyngeal carcinoma (NPC) cell line was examined. The levels of S100P protein in NPC tissues were analyzed using immunohistochemistry, and small interfering RNA silenced S100P expression in C666-1 cells. Subsequently, cell proliferation, colony formation, migration and wound-healing assays were performed in order to assess whether the knockdown of S100P was able to influence the biological behavior of C666-1 cells. The expression levels of the receptor for advanced glycation end products (RAGE) were analyzed using a western blot following the inhibition of S100P. The immunohistochemistry results revealed that S100P was elevated in expression in 45/78 (57.7%) of patients with NPC, as compared with 5/30 (16.7%) of patients with benign inflammation. The S100P protein levels correlated with the rates of proliferation and migration in C666-1 cells. Additionally, reduced S100P expression levels altered a series of intracellular events, including the downregulation of epidermal growth factor receptor, cluster of differentiation (CD) 44, matrix metalloproteinase (MMP) 2 and MMP9 protein expression. In addition, RAGE expression was downregulated in the S100P silenced C666-1 cells, as detected by western blot analysis. These data suggest that S100P is important during the development and progression of nasopharyngeal cancer. Therefore, S100P may provide a novel treatment target for NPC.
Keywords: S100 calcium binding protein P, proliferation, migration, C666-1 cell line, nasopharyngeal carcinoma
Introduction
The S100 calcium binding proteins are a multi-gene family composed of ≥20 members in humans, each encoded by a separate gene (1). The deregulated expression of several members of the S100 protein family, including S100 calcium binding protein (S100) B, S100A2, S100A4, S100A6, S100A11 and S100P has been reported to be associated with the progression and metastasis of various types of human cancer (2). Head and neck cancer is eighth leading cause of cancer-associated mortality worldwide (3). Previous studies have indicated the potential association between S100 proteins and head and neck cancer (4,5), and the altered expression of various S100 proteins has been reported in primary and metastatic laryngeal carcinoma (6). In a previous study, S100A11 was revealed to be overexpressed in laryngeal tumor tissues, as compared with the corresponding noncancerous tissues, which was determined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis (7). In addition, S100A11 expression was correlated with human epithelial type 2 cell migration (7). The expression of S100A4 has also been correlated with invasion and metastasis in oral squamous cell carcinoma (8).
Nasopharyngeal carcinoma (NPC) is the most prevalent head and neck cancer in Southern China and Southeast Asia (9). NPC is distinct from other types of head and neck cancer due to its sensitivity to radiotherapy and chemotherapy (10). Although radiotherapy enables a high cure rate for early-stage NPC, the treatment outcome for the advanced stage disease remains poor (11). Local recurrences and the high frequency of distant metastasis are two major causes of treatment failure in NPC following radiotherapy (12).
S100P is a 95 amino acid protein and a member of the S100 family of EF-hand motif calcium-binding proteins, which was initially purified from the human placenta and exhibits a restricted cellular distribution (13). Its deregulated expression has previously been observed in association with the progression and metastasis of various types of human cancer, including breast (14), lung (15), gastric (16) and prostate cancer (17). Despite these observations, the functional role and the underlying mechanism of action of S100P in nasopharyngeal carcinoma requires further investigation.
In the present study, the differential expression of S100P in NPC tissues was examined using immunohistochemistry (IHC), and the changes of proliferation and migration of C666-1 cells were analyzed following S100P silencing with the aim of investigating the role of S100P in NPC.
Patients and methods
Patients and tissue samples
Pathological tissue specimens were obtained retrospectively from 78 patients with nasopharyngeal carcinoma and 30 patients with benign inflammation at The Department of Otolaryngology-Head and Neck Surgery, Changzheng Hospital, Second Military Medical University (Shanghai, China) between January 2005 and June 2008. Patient characteristics were obtained from the medical records (Table I). The median age of the 78 patients with nasopharyngeal carcinoma was 51.78 years old (range, 21–89 years), and 22 (28.21%) of the patients were >60 years of age. The median age of 30 patients with benign inflammation was 52.09 years (range, 19–78 years), and 8 (26.67%) of the patients were >60 years of age. There were 53 males (67.95%) and 25 females (32.05%) with nasopharyngeal carcinoma, compared with 22 males (73.33%) and 8 females (26.67%) with benign inflammation.
Table I.
Clinical characteristics of the patients with nasopharyngeal carcinoma.
| S100Ps | |||
|---|---|---|---|
| Patient characteristics | Negative | Positive | P-value |
| Sex | 0.835 | ||
| Male | 22 | 31 | |
| Female | 11 | 14 | |
| Age, years | 0.234 | ||
| <60 | 20 | 33 | |
| ≥60 | 13 | 12 | |
| Primary tumor | 0.066 | ||
| T1 | 22 | 38 | |
| T2-T4 | 11 | 7 | |
| Nodal status | 0.656 | ||
| N0-N1 | 20 | 25 | |
| N2-N3 | 13 | 20 | |
| Stage | 0.929 | ||
| I–II | 18 | 25 | |
| III–IV | 15 | 20 | |
| Histological grade | 0.392 | ||
| Keratinized | 5 | 3 | |
| Non-keratinized | 8 | 15 | |
| Undifferentiated | 20 | 27 | |
S100P, S100P calcium binding protein P; T, tumor; N, node.
The pathological tissue specimens of all the patients who had undergone neither chemotherapy nor radiotherapy underwent biopsy for immunohistochemical examination of S100P expression levels. The patients follow-up data was available for >5 years. All the tissue specimens were obtained for the present study following the receipt of written informed patient consent. The collection of these tissue samples was undertaken with the approval of The Second Military Medical University Ethics Committee.
IHC
IHC staining was performed using a Dako EnVision Peroxidase/DAB system (Dako North America, Inc., Carpinteria, CA, USA) according to the manufacturer's protocol. The tissue samples from each patient were fixed in formalin and embedded in paraffin. Subsequently, the tissue sections (4 µm thick) were deparaffinized using xylene, rehydrated with graded ethanol and heated for 1 h at 65°C. The sections were submerged in EDTA antigenic retrieval buffer (pH 8.0, K800221-2, EnVision FLEX+, Agilent Tech) and were heated with high heat for 3 min, then heated with low heat for 15 min using a microwave (Galanz microwave oven P70D20P-N9). Following treatment with 0.3% H2O2 for 15 min at room temperature to block endogenous peroxidase activity, the tissue sections were treated with 1% bovine serum albumin (ab192603; Abcam, Cambridge, UK) for 30 min in the 37°C incubator to reduce non-specific binding, and subsequently incubated with a primary rabbit polyclonal anti-S100P antibody (dilution, 1:500; ab124743; Abcam) overnight at 4°C, followed by incubation with horseradish peroxidase-labeled secondary antibodies (dilution, 1:300; ab181875; Abcam) at 4°C for 30 min. The slides were developed using diaminobenzidine tetrahydrochloride and counterstained with hematoxylin. Immunohistochemically stained tissue sections were scored independently by two pathologists (Department of Pathology, Changzheng Hospital, Second Military Medical University) who were blinded to the clinical parameters. The tissue specimens were defined as positive for S100P expression if the tumor cells were distinctly stained by the anti-S100P antibody. Entire tissue sections were imaged under a microscope to enable the scoring of S100P staining. In each section, 5 fields were selected randomly under a high power microscope (magnification, ×400), within each field 200 tumor cells were counted. The staining intensity was scored as follows: 0 (negative), 1 (weak) and 2 (intense). The extent of staining was scored as 0 (0–10%), 1 (11–25%), 2 (26–50%) and 3 (51–100%), according to the proportion of positive-staining areas in the entire carcinoma area, or the entire section for the benign tissue samples. The product of the intensity and the extent of staining scores was used as the final staining score (0–6) for S100P (score=intensity of staining score × extent of staining score). For the purpose of statistical evaluation, tumors (nasopharyngeal carcinoma tissue sections) with a final staining score of >2 were considered to be positive.
Cell culture and maintenance of cell lines
The C666-1 cell line was obtained from the cell bank of The Central Laboratory of Xiangya Hospital (Central South University, Changsha, China) (18). C666-1 cells were cultured in RPMI-1640 (GE Healthcare Life Sciences; Hyclone, Logan, UT, USA) supplemented with 12% fetal bovine serum (FBS) (Biowest, Maine et Loire, France) and 1% penicillin and streptomycin (P/S) (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and were maintained in a humidified 37°C incubator containing 5% CO2.
Transfection of small interfering RNA (siRNA)
S100P knockdown experiments were performed by transfecting S100P siRNA into C666-1 cells. siRNA against human S100P was chemically synthesized by GenePharma Co., Ltd., (Shanghai, China) according to the sequence published previously by Arumugam et al (19). The siRNA sequences were as follows: Forward, 5′-AAUGGAGAUGCCCAGGUGGACTT-3′ and reverse, 5′-GUCCACCUGGGCAUCUCCAUUTT-3′. The negative control siRNA sequences were as follows: Forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′. S100P siRNA transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. C666-1 cells were plated at a density of 2×105 cells/well in 6-well plates and were incubated at 37°C overnight in 2 ml RPMI-1640 (GE Healthcare Life Sciences; Hyclone) supplemented with 12% fetal bovine serum (FBS; Biowest) without antibiotics. Upon reaching 30–50% confluency, the culture medium was aspirated. A total of 5 µl of 20 µM siRNA and 5 µl Lipofectamine® 2000 were combined for 20 min and subsequently added to a final volume of 2 ml of serum-free Gibco™ Opti-MEM™ medium (Thermo Fisher Scientific, Inc.). The medium was replaced 5 h following transfection, and RNA and protein were harvested from the cells at 24 or 48 h post-transfection for the evaluation of S100P knockdown. The effects of S100P gene knockdown were examined using RT-qPCR and western blotting.
RNA isolation and RT-qPCR analysis
Total RNA (2 µg) was extracted from the S100P siRNA-transfected C666-1 cells using TRIzol® reagent (Ambion; Thermo Fisher Scientific, Inc.), and reverse transcription was performed using the RevertAid First Strand cDNA Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.). The primers were chemically synthesized by GenePharma. The primer sequences were as follows: Forward, 5′-ATGACGGAACTAGAGACAGCCATGGGC-3′ and reverse, 5′-GGAATCTGTGACATCTCCAGCGCATCA-3′ (19). The primer sequences for the internal control GAPDH were as follows: Forward, 5′-GGTGGTCTCCTCTGACTTCAACA-3′ and reverse, 5′-GTTGCTGTAGCCAAATTCGTTGT-3′. The reaction mixtures were incubated for 60 min at 42°C followed by heating at 72°C for 5 min for reverse transcription. The resulting cDNA (2 µg) was added to a 20 µl reaction mix containing 0.8 µl primers and 10 µl SYBR®-Green Real Time PCR Master mix (Toyobo Co., Ltd., Osaka, Japan). RT-qPCR was performed using the ABI 7300 system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were set as follows: 40 cycles at 95°C for 3 min, 95°C for 15 sec and 60°C for 31 sec. The expression value of S100P compared with that of GAPDH was determined using the 2−ΔΔCq method (20).
Western blotting
The cell lysates were prepared using radioimmunoprecipitation assay buffer supplemented with phenylmethylsulfonyl fluoride (Beyotime Institute of Biotechnology, Shanghai, China), followed by centrifugation at 12,000 × g for 5 min at 4°C. Protein concentrations were estimated using the bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology), according to the manufacturer's protocol; 50 µg protein was denatured in 1X loading buffer (Beyotime Institute of Biotechnology) at 98°C for 5 min and separated by 12% SDS-PAGE, and then electrotransferred onto nitrocellulose membranes with an electroblotting apparatus (Bio-Rad Laboratories Inc., Hercules, CA, USA). Western blot analysis was conducted using the primary anti-human S100P antibodies (dilution, 1:1,000; Abcam; ab124743), receptor for advanced glycation end products (RAGE; dilution, 1:1,000; Abcam; ab3611), epidermal growth factor receptor (EGFR; dilution, 1:1,000; 18986-1-AP; ProteinTech Group Inc., Chicago, IL, USA), cluster of differentiation (CD)44 (dilution, 1:1,000; 15675-1-AP; ProteinTech Group Inc.), matrix metalloproteinase (MMP)2 (dilution, 1:1,000; 10373-2-AP; ProteinTech Group Inc.) and MMP9 (dilution, 1:1,000; 10375-2-AP; ProteinTech Group Inc.). The membranes were incubated overnight at 4°C with primary antibodies. The secondary antibodies [Anti-rabbit IgG (heavy chain or light chain binding)] goat; dilution, 1:10,000; 611-1302) conjugated with horseradish peroxidase were incubated with the nitrocellulose membrane at room temperature for 1 h. Subsequently, the membranes were washed with Tris-buffered saline and Tween-20 and treated with an enhanced chemiluminescence reagent (EMD Millipore, Billerica, MA, USA). Immunoreactive bands were detected by exposure to X-ray film. For quantification, the target protein was normalized to the internal standard protein GAPDH through comparison of the gray scale values. This analysis was performed using Gel-Pro Analyzer software, version 4.0 (Media Cybernetics, Inc., Rockville, MD, USA).
Cell proliferation assay
The proliferation of C666-1 cells transfected with S100P siRNA or negative control siRNA was determined using a Cell Counting kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Triplicates of cell samples were seeded into 96-well plates at a density of 5×103 cells/well. At the indicated time points (24, 48 and 72 h), 10 µl CCK-8 solution was added to each well. Following incubation for 2 h at 37°C, the absorbance was measured at 450 nm using a spectrophotometer (Thermo Fisher Scientific, Inc.).
Colony formation assay
The C666-1 cells transfected with S100P siRNA or negative control siRNA were seeded at a density of 1,000 cells/well in a 6-well plate. The cells were cultured in 2 ml RPMI-1640 supplemented with 12% FBS and 1% P/S, and were incubated at 37°C and 5% CO2 for 2 weeks. The culture medium was replaced every three days. Upon completion of the incubation, the culture medium was aspirated and the cells were washed with PBS, fixed in 1 ml methanol for 30 min at room temperature and stained with 0.4% crystal violet for 20 min at room temperature. The number of colonies containing >50 cells was counted manually using an inverted microscope (Olympus Corp., Tokyo, Japan) and averaged from the duplicate wells.
Migration assay
The migration of the C666-1 cells transfected with S100 siRNA or negative control siRNA was assessed using Transwell cell culture chamber inserts (Corning Incorporated, Corning, NY, USA) with an 8-µm pore size. A total of 1×104 cells that had been treated with S100P siRNA or negative control siRNA for 24 h were seeded into Transwell filter membrane chambers with 100 µl RPMI-1640 supplemented with 1% FBS. A total of 600 µl RPMI-1640 supplemented with 20% FBS was added to the lower compartment as a chemoattractant. Following incubation for 20 h at 37°C in a humidified 5% CO2 atmosphere, the cells on the upper surfaces of the wells were removed with cotton swabs and the cells on the upper surfaces that had migrated to the lower chamber, were fixed in cold methanol at 4°C for 10 min, and stained with 0.4% crystal violet for 10 min at room temperature. Excess stain was removed using physiological saline and the chambers were air-dried at room temperature for ~20 min. For each experiment, five fields on the undersides of the membranes were randomly selected for imaging, and the transmigrated cells in the five random fields were counted and the mean was determined from the duplicate wells.
Wound healing assay
C666-1 cells transfected with S100P siRNA or negative control siRNA for 24 h were seeded at a density of 2×106 single cells per well in a 6-well plate at 90–95% confluency. The cell monolayer was scratched using a sterile 200 µl pipette tip. The cells were rinsed with fresh medium [(RPMI-1640; GE Healthcare Life Sciences; Hyclone) supplemented with 5% FBS (Biowest)] to remove any free-floating cells and debris. Images were captured immediately using an inverted microscope equipped with a digital camera (Olympus) and wound healing was observed at various time points (0, 12, 24 and 48 h) around the scrape line. Images of representative scrape lines were captured. The wound area was measured using Adobe Photoshop software version 7.0 (Adobe Systems, Inc., San Jose, CA, USA).
Statistical analysis
The data were analyzed using SPSS version 18.0 (SPSS, Inc., Chicago, IL, USA). χ2 test and Yates' correction were used to analyze the correlation between S100P expression levels and the histological type, stage, age and gender distribution. Kaplan-Meier analysis was used to analyze the correlation between S100P immunoreactivity and overall and disease-free survival. All the experiments were performed ≥3 times and the results are presented as the mean ± standard deviation. Statistical comparisons between two groups of data were performed using the two-tailed Student's t-test. Multiple group comparisons were performed using one-way analysis of variance. P<0.05 was considered to indicate a statistically significant difference.
Results
The clinical and pathological significance of S100P expression levels in nasopharyngeal cancer
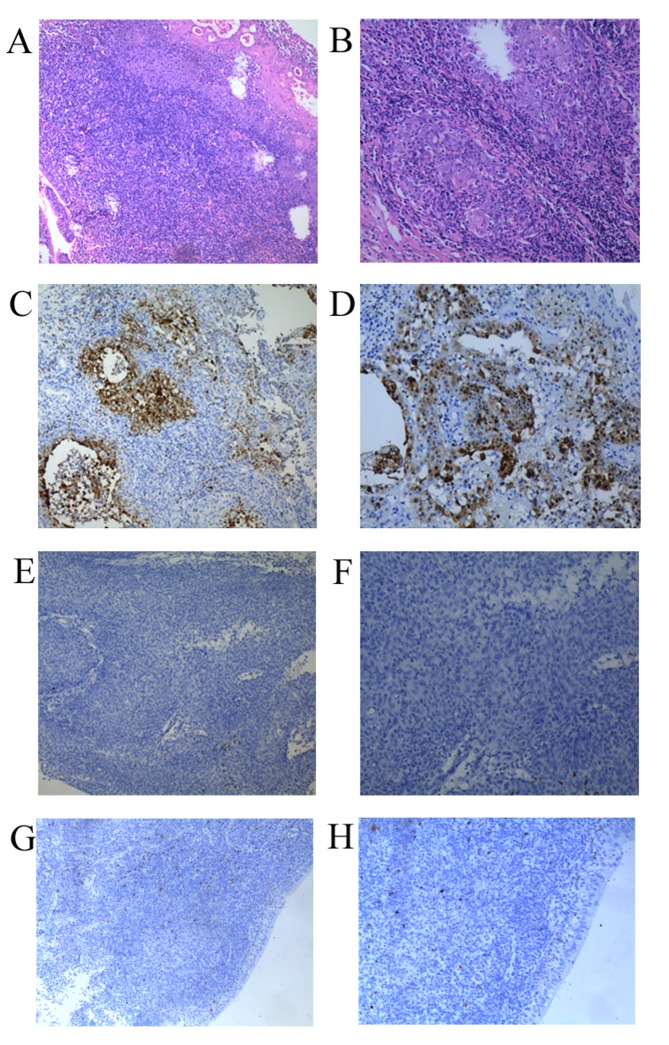
To investigate the clinical and pathological significance of S100P expression levels in nasopharyngeal cancer, the tissues of 78 patients with NPC and 30 patients with benign inflammation were analyzed using IHC. Intense positive nuclear staining was observed in all the neoplastic cells in the positive controls. The tissue specimens from the patients with NPC exhibited diffuse or focal positive nuclear staining. Fig. 1A and B present the typical nasopharyngeal carcinoma cells. Immunoreactivity was more pronounced in the basal layer of the normal and dysplastic epithelium (Fig. 1C, D, E and F). Of a total of 78 NPC samples, positive staining for the S100P protein was observed in 45 (57.7%) tissue samples, whereas no staining was detected in 33 (42.3%) tissue samples. No S100P staining was observed in the 30 benign inflammation tissues (Fig. 1G and H). The correlation between S100P expression levels and the histological type, stage, age and sex distribution was examined (Table I), and no significant differences were observed according to the χ2 test and Yates' correction (P>0.05). The median follow-up time was 63.7 months (range, 13–93 months), and the 5-year cumulative survival rate was 68.3±2.5% standard error. According to Kaplan-Meier analysis, a statistically significant correlation was observed between S100P immunoreactivity and overall and disease-free survival (Fig. 2; P<0.01).
Figure 1.
Hematoxylin and eosin staining in NPC tissues. (A) S100P positive staining in NPC (magnification, ×100). (B) S100P positive staining in NPC (magnification, ×200). (C) S100P negative staining in NPC (magnification, ×100). (D) S100P negative staining in NPC (magnification, ×200). (E) S100P negative staining in benign inflammation (magnification, ×100). (F) S100P negative staining in benign inflammation (magnification, ×200). (G) magnification, ×100; (H) magnification, ×200). NPC, nasopharyngeal carcinoma; S100P, S100 calcium binding protein P.
Figure 2.
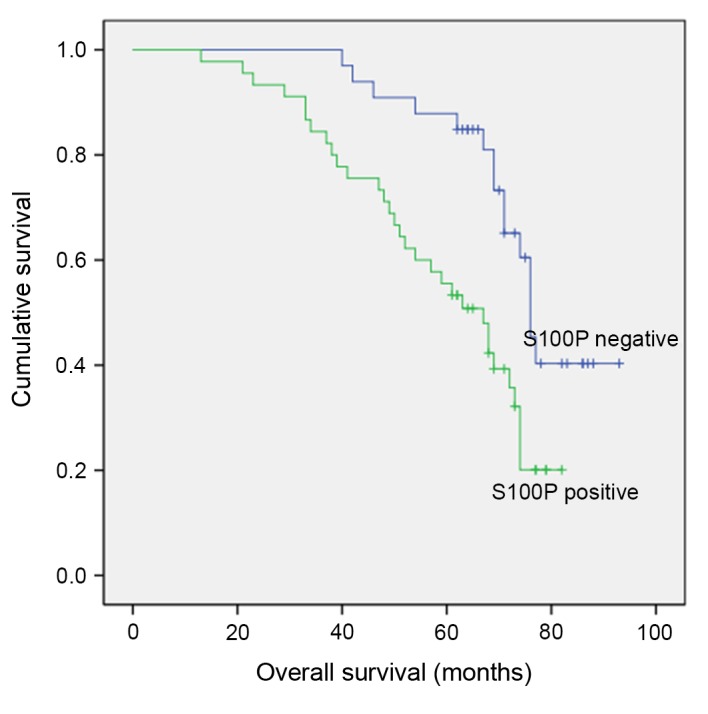
Statistically significant association was observed between S100P immunoreactivity and patient overall survival. (P<0.01) S100P, S100 calcium binding protein P.
Knockdown of S100P decreases cell proliferation and invasion in C666-1 cells
In siRNA S100P transfected C666-1 cells, decreased levels of S100P mRNA and protein were observed using RT-qPCR and western blot analyses, respectively (P<0.001; Fig. 3). Cell growth, colony formation and migration assays were performed. S100P knockdown produced no significant effects on cell proliferation at 24 (P<0.622) and 48 h (P<0.245); however, cell proliferation was significantly decreased at 72 h (P<0.001) in the S100P siRNA group, compared with the negative control siRNA group and the untreated control group (Fig. 4A). S100P knockdown reduced the colony forming ability of the S100P siRNA transfected cells (Fig. 4B and C). Additionally, the effects on C666-1 cell migration were assessed by Transwell and wound healing assays, revealing that the knockdown of S100P significantly reduced the invasive ability of cells (P<0.001; Fig. 5A and B). As presented in Fig. 5C and D (P<0.001), photomicrographs captured at 48 h following wounding of the cell monolayer revealed delayed wound closure in cells transfected with S100P siRNA, compared with cells transfected with negative control siRNA and the untreated controls. The EGFR, CD44, MMP2 and MMP9 proteins are important during cancer cell invasion and metastasis (21–23). Therefore, the protein expression levels of EGFR, CD44, MMP2 and MMP9 in C666-1 cells that had been treated with S100P or negative control siRNA were assessed using western blotting. The results revealed that the expression levels of these proteins were decreased in the S100P siRNA group, compared with the negative control siRNA and untreated control groups (Fig. 6; EGFR, P<0.01; CD44, P<0.01; MMP2, P<0.001; MMP9, P<0.01). Taken together, these results indicate that a reduction in the level of S100P expression is able to suppress the growth and invasion of C666-1 cells. These functions support our hypothesis that the regulation of S100P expression is an important step by which cancer cells are able to promote their growth and migration during carcinogenesis.
Figure 3.
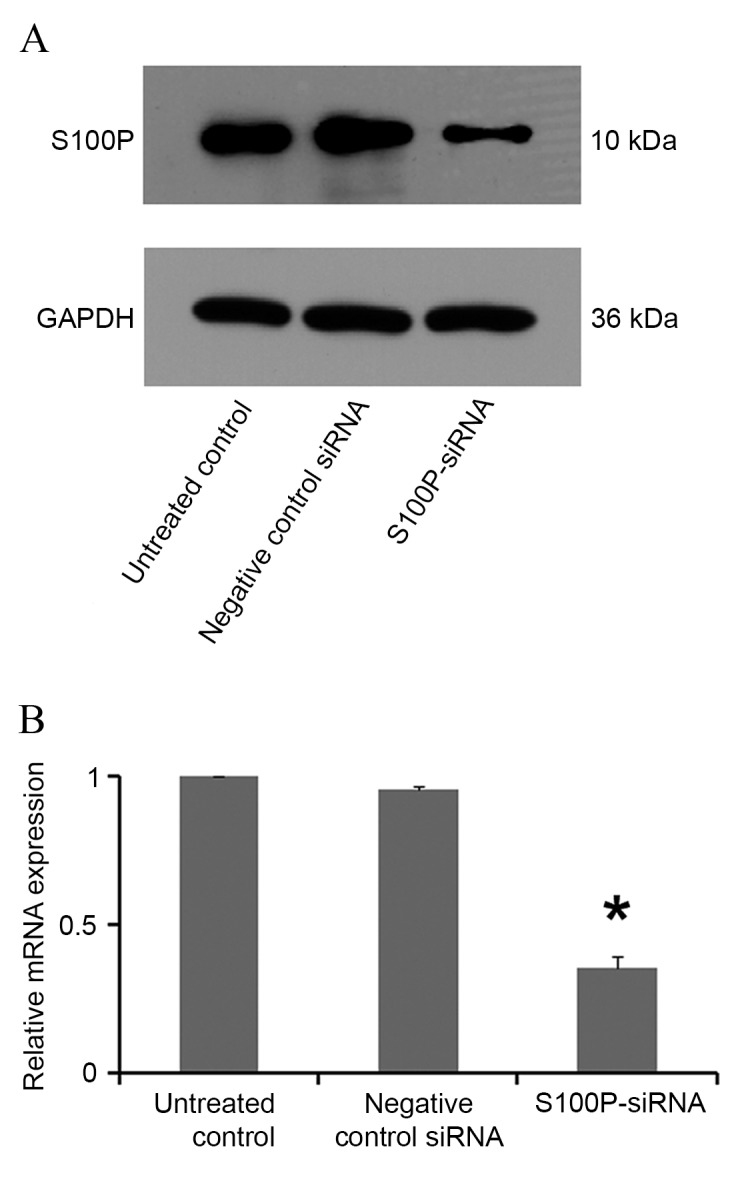
Effect of siRNA-mediated S100P knockdown on C666-1 cells at 48 h post transfection. (A) Western blotting and (B) RT-qPCR analyses of S100P protein and mRNA expression levels, respectively, in S100P-siRNA-transfected C666-1 cells. siRNA, small interfering RNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; S100P, S100 calcium binding protein P. *P<0.01.
Figure 4.
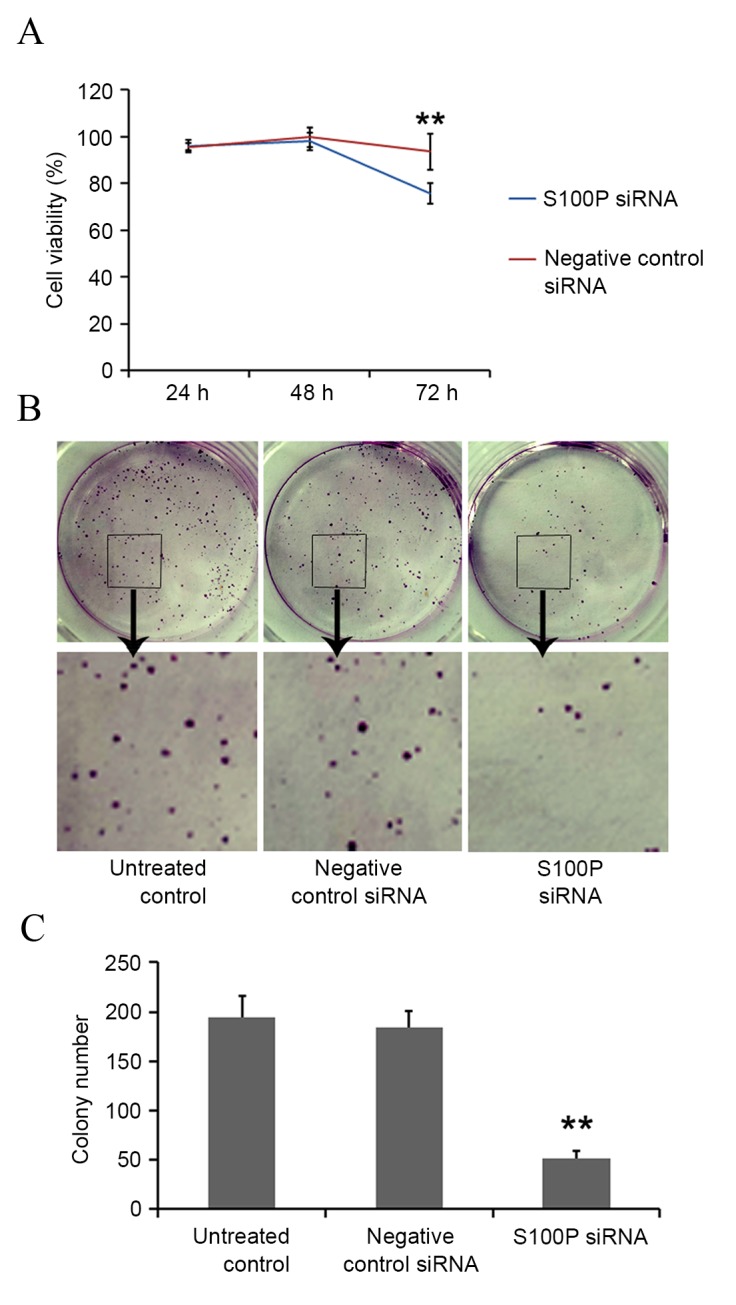
Downregulation of S100P expression inhibits C666-1 cell proliferation. (A) Cell counting kit-8 assay indicated that cell viability was significantly decreased at 72 h in the S100P siRNA group, compared with the negative control siRNA group and the untreated control group (P<0.001). (B) Colony formation assays indicated that the colony numbers were decreased in the S100P siRNA group compared with the negative control siRNA and untreated control groups. Images of colonies in six-well plates were taken. (C) The numbers of colonies were counted. (P<0.01). **P<0.01. siRNA, small interfering RNA; S100P, S100 calcium binding protein P.
Figure 5.
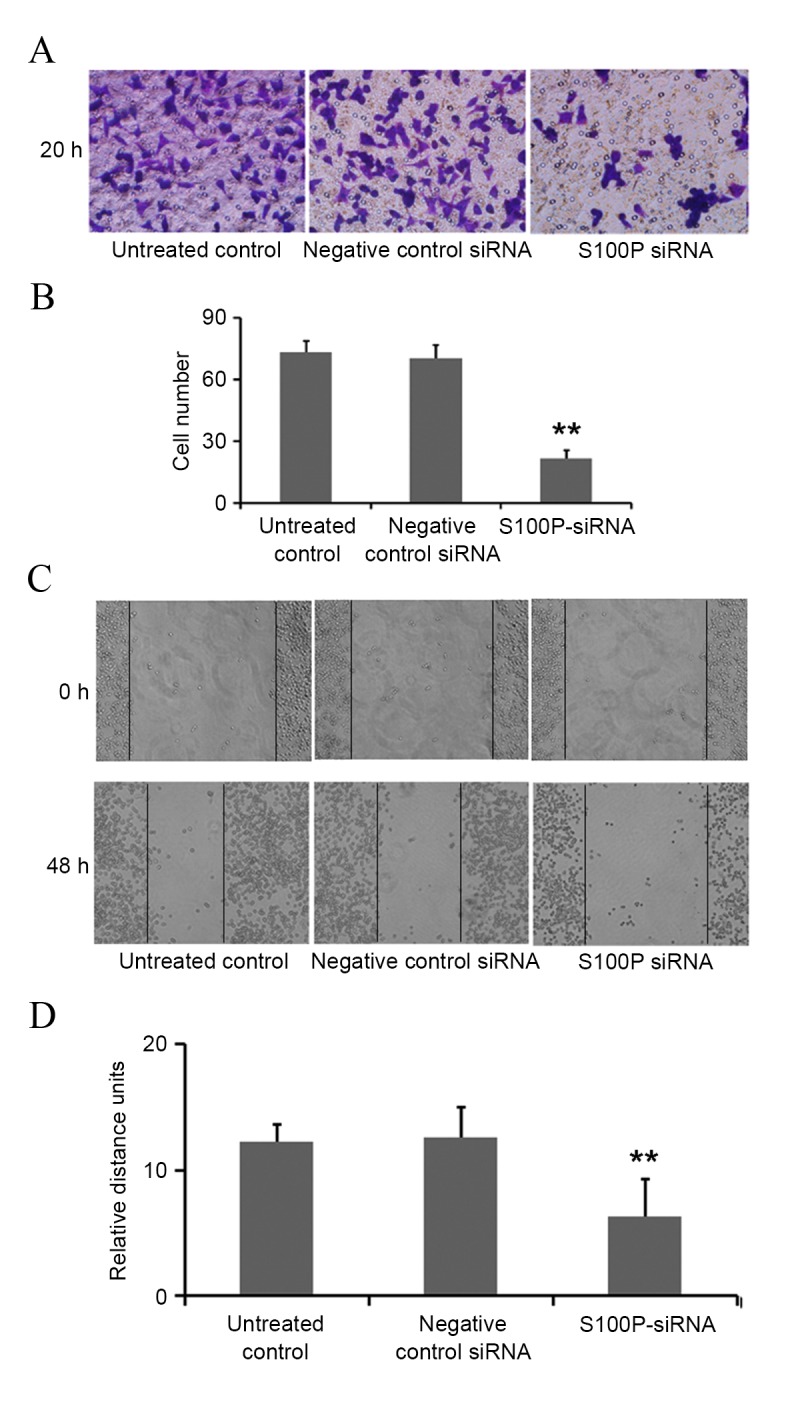
Downregulation of S100P expression inhibits C666-1 cell migration. (A) Sample images showing the results of the Transwell assay that the cells in the S100P-siRNA group that had migrated to the lower Transwell membrane surfaces was reduced compared with the negative control siRNA and untreated control groups. (B) Graphs indicated the average number of cells from five random fields in the three groups (P<0.01). (C) The migration rate was quantified by evaluating the distance between the edges of the scratch in a scratch migration assay. (D) Graphs indicated that the cells in the S100P-siRNA group migrated slowly, compared with those in the negative control siRNA and untreated control groups (P<0.01). **P<0.01. S100P, S100 calcium binding protein P; siRNA, small interfering RNA.
Figure 6.
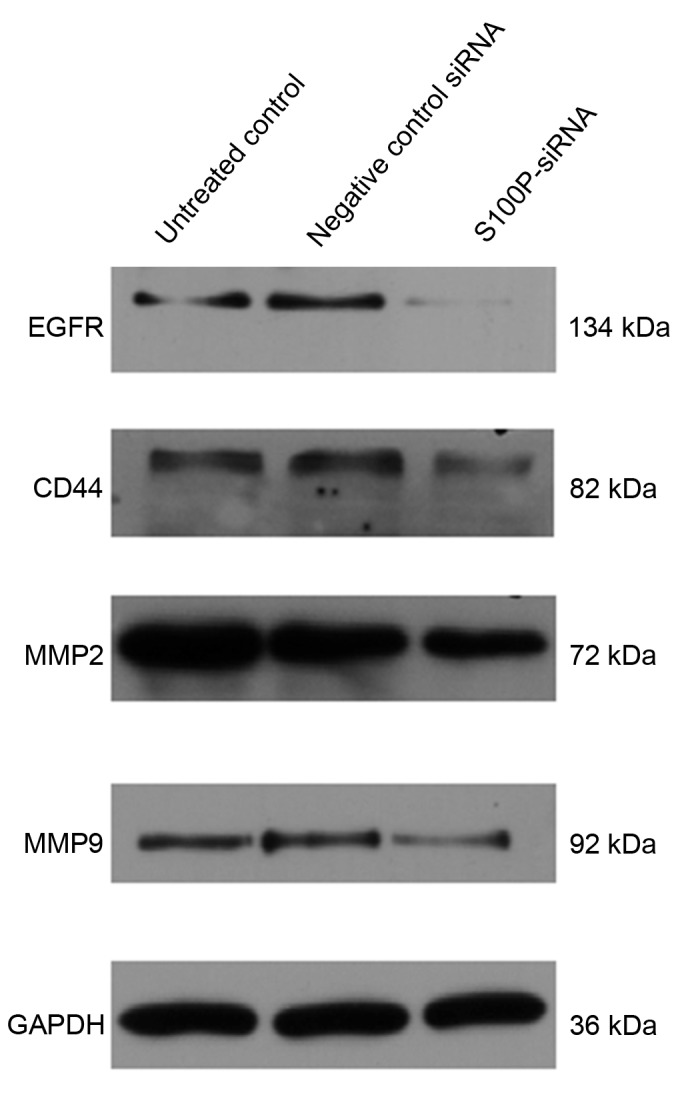
Effects of S100P silencing on the expression levels of migration-associated proteins in C666-1 cells. Western blotting was used to analyze the protein expression levels of EGFR, CD44, MMP2 and MMP9 in C666-1 cells that had been transfected with S100P siRNA. The expression levels of these proteins were decreased in the S100P siRNA group compared with the negative control siRNA and untreated control groups. S100P, S100 calcium binding protein P; siRNA, small interfering ribonucleic acid; EGFR, epidermal growth factor receptor; CD44, cluster of differentiation 44; MMP, matrix metalloproteinase; MMP, matrix metalloproteinase.
Extracellular S100 proteins act via a variety of cellular receptors (24,25) and RAGE has been suggested to be a general receptor for the S100 family of proteins (26). Using western blot analysis, the present study observed that RAGE expression was downregulated in the S100P siRNA group, compared with the negative siRNA control and untreated control groups (Fig. 7; P<0.001).
Figure 7.
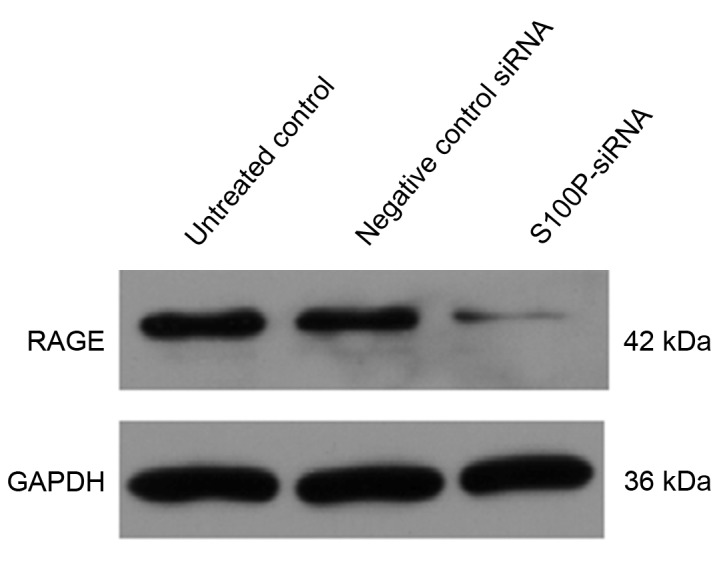
RAGE protein expression levels were analyzed using western blotting in C666-1 cells that had been transfected with S100P siRNA. RAGE expression was decreased in the S100P siRNA group compared with the negative siRNA control and untreated control groups. RAGE, receptor for advanced glycation end products; S100P, S100 calcium binding protein P.
Discussion
The overexpression of S100P, which was first purified from the human placenta, has been detected in a variety of types of cancer, and correlates with tumor growth and metastasis (19,27). Several studies have linked S100P expression levels to cell proliferation, invasion and migration in various forms of cancer (28).
To investigate the clinical and pathological significance of S100P expression in nasopharyngeal cancer, the expression levels of the S100P protein in human nasopharyngeal tissues were evaluated using IHC. The results revealed significantly increased S100P expression levels in 45/78 (57.7%) patients with NPC. In addition, a significant correlation between S100P immunoreactivity and overall disease-free survival was observed.
A number of in vitro and in vivo studies have evaluated the effects of the silencing of genes involved in certain signaling pathways that promote cancer progression, including oncogenesis, apoptosis, cell cycle regulation, cell senescence, tumor-host interactions and resistance to conventional therapies (29,30). In the present study, the potential effects of S100P on the characteristics of nasopharyngeal cancer were investigated, and siRNA was used to silence S100P expression in C666-1 cells. The results revealed that S100P protein and mRNA expression levels were significantly reduced in the S100P siRNA-transfected group. Cell growth assays were performed to evaluate the association between S100P expression levels and C666-1 cell proliferation. The results from the CCK-8 and colony formation assays revealed that S100P silencing decreased the proliferative ability of C666-1 cells, which suggests that S100P may be a tumor growth-associated gene in NPC.
Cancer metastasis is a multistep process involving migration and invasion through the tumor stroma, intravasation, tumor cell dissemination, extravasation and cell growth at the metastatic sites (31). The present study investigated the migratory characteristics of C666-1 cells, following S100P silencing, using Transwell and wound-healing assays. The migration of C666-1 cells was observed to be significantly reduced when S100P expression was inhibited by siRNA. Furthermore, the expression levels of EGFR, CD44, MMP2 and MMP9 were decreased significantly when S100P was silenced, suggesting that S100P is associated with cell migration in NPC. EGFR is a type I receptor tyrosine kinase that is overexpressed in a number of solid tumors, including types of head and neck carcinoma, and is associated with a poor prognosis following treatment (32). CD44 is a cell-surface molecule that has been implicated in a diverse range of cell-cell and cell-matrix interactions (33). MMPs, a family of zinc-binding endopeptidases, possess an established association with cancer-cell invasion and metastasis (34). EGFR, CD44 and MMPs are therefore key factors during cancer development, progression and metastasis, and downregulation of the expression of these factors, as a result of decreased S100P expression levels, suggests a correlation between S100P expression levels and C666-1 cell proliferation and migration (35).
RAGE, a member of the immunoglobulin protein family of cell surface molecules (36), shares structural homology with other immunoglobulin-like receptors (37) and is important in certain human pathologies, including cancer (38). The present study analyzed the expression levels of RAGE protein in S100P siRNA transfected C666-1 cells using western blot analysis. The results revealed that RAGE expression was downregulated in the S100P siRNA group, as compared with the negative siRNA control and untreated control groups. A number of S100 proteins interact with RAGE in vitro and trigger RAGE-dependent signaling in cell-based assays. For instance, Arumugam et al (39) demonstrated that S100P is able to trigger the activation of nuclear factor-κB via the mitogen activated protein kinase signaling pathway in a RAGE-dependent manner in BxPC3 and SW480 adenocarcinoma cells. The present study revealed that the siRNA-mediated knockdown of S100P expression in C666-1 cells significantly inhibits cancer cell growth, migration and invasion in vitro. Further studies may investigate the role of S100P by examining its effects on tumor growth and metastasis in vivo.
In conclusion, the results of the present study suggested that S100P may be an important regulatory protein during the promotion of NPC cell proliferation and migration. Additionally, the current study provided information about the function of proliferation and migration of S100P in C666-1 cells that may aid the elucidation of the molecular mechanisms underlying tumor metastasis and the identification of clinically relevant biomarkers for metastasis prevention in NPC.
Acknowledgements
The present study was supported by the Basic Research Project of The Shanghai Technology Commission (grant no. 12JC1411100), The Key Disciplines Group Construction Project of the Pudong Health Bureau of Shanghai (grant no. PWZxq 2014–09) and The Project of The Shanghai Technology Commission (grant nos. 14DZ1940103 and 14411960400).
References
- 1.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of thenomenclature) Biochem Biophys Res Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 2.Heizmann CW, Fritz G, Schäfer BW. S100 proteins: Structure, functions and pathology. Front Biosci. 2002;7:d1356–d1368. doi: 10.2741/A846. [DOI] [PubMed] [Google Scholar]
- 3.Ragin CC, Modugno F, Gollin SM. The epidemiology and risk factors of head and neck cancer: A focus on human papillomavirus. J Dent Res. 2007;86:104–114. doi: 10.1177/154405910708600202. [DOI] [PubMed] [Google Scholar]
- 4.Zha C, Jiang XH, Peng SF. iTRAQ-based quantitative proteomic analysis on S100 calcium binding protein A2 in metastasis of laryngeal cancer. PLoS One. 2015;10:e0122322. doi: 10.1371/journal.pone.0122322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moriyama-Kita M, Endo Y, Yonemura Y, Heizmann CW, Schäfer BW, Sasaki T, Yamamoto E. Correlation of S100A4 expression with invasion and metastasis in oral squamous cell carcinoma. Oral Oncol. 2004;40:496–500. doi: 10.1016/j.oraloncology.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Nakashima T, Yano G, Hayashi I, Katsuta Y. Epithelial membrane antigen and S-100 protein-labeled cells in primary and metastatic laryngeal carcinomas. Head Neck. 1992;14:445–451. doi: 10.1002/hed.2880140604. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Zhang Z, Li L, Zhang J, Wang J, Fan J, Jiao B, Zhao S. S100A11 is a migration-related protein in laryngeal squamous cell carcinoma. Int J Med Sci. 2013;10:1552–1559. doi: 10.7150/ijms.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moriyama-Kita M, Endo Y, Yonemura Y, Heizmann CW, Schäfer BW, Sasaki T, Yamamoto E. Correlation of S100A4 expression with invasion and metastasis in oral squamous cell carcinoma. Oral Oncol. 2004;401:496–500. doi: 10.1016/j.oraloncology.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Sung FL, Poon TC, Hui EP, Ma BB, Liong E, To KF, Huang DP, Chan AT. Antitumor effect and enhancement of cytotoxic drug activity by cetuximab in nasopharyngeal carcinoma cells. In Vivo. 2005;19:237–245. [PubMed] [Google Scholar]
- 10.Paiar F, Di Cataldo V, Zei G, Pasquetti EM, Cecchini S, Meattini I, Mangoni M, Agresti B, Iermano C, Bonomo P, Biti G. Role of chemotherapy in nasopharyngeal carcinoma. Oncol Rev. 2012;6:e1. doi: 10.4081/oncol.2012.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu H, Peng L, Yuan X, Hao Y, Lu Z, Chen J, Cheng J, Deng S, Gu J, Pang Q, Qin J. Concurrent chemoradiotherapy in locally advanced nasopharyngeal carcinoma: A treatment paradigm also applicable to patients in Southeast Asia. Cancer Treat Rev. 2009;35:345–353. doi: 10.1016/j.ctrv.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Baujat B, Audry H, Bourhis J, Chan AT, Onat H, Chua DT, Kwong DL, Al-Sarraf M, Chi KH, Hareyama M, et al. Chemotherapy in locally advanced nasopharyngeal carcinoma: An individual patient data meta-analysis of eight randomized trials and 1,753 patients. Int J Radiat Oncol Biol Phys. 2006;64:47–56. doi: 10.1016/j.ijrobp.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 13.Becker T, Gerke V, Kube E, Weber K. S100P, a novel Ca(2+)-binding protein from human placenta. cDNA cloning, recombinant protein expression and Ca2+ binding properties. Eur J Biochem. 1992;207:541–547. doi: 10.1111/j.1432-1033.1992.tb17080.x. [DOI] [PubMed] [Google Scholar]
- 14.Da Silva ID Guerreiro, Hu YF, Russo IH, Ao X, Salicioni AM, Yang X, Russo J. S100P calcium-binding protein overexpression is associated with immortalization of human breast epithelial cells in vitro and early stages of breast cancer development in vivo. Int J Oncol. 2000;16:231–240. [PubMed] [Google Scholar]
- 15.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 16.Shyu RY, Huang SL, Jiang SY. Retinoic acid increases expression of the calcium-binding protein S100P in human gastric cancer cells. J Biomed Sci. 2003;10:313–319. doi: 10.1007/BF02256450. [DOI] [PubMed] [Google Scholar]
- 17.Basu GD, Azorsa DO, Kiefer JA, Rojas AM, Tuzmen S, Barrett MT, Trent JM, Kallioniemi O, Mousses S. Functional evidence implicating S100P in prostate cancer progression. Int J Cancer. 2008;123:330–339. doi: 10.1002/ijc.23447. [DOI] [PubMed] [Google Scholar]
- 18.Cheung ST, Huang DP, Hui AB, Lo KW, Ko CW, Tsang YS, Wong N, Whitney BM, Lee JC. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein-Barr virus. Int J Cancer. 1999;83:121–126. doi: 10.1002/(SICI)1097-0215(19990924)83:1<121::AID-IJC21>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 19.Arumugam T, Simeone DM, Van Golen K, Logsdon CD. S100P promotes pancreatic cancer growth, survival, and invasion. Clin Cancer Res. 2005;11:5356–5364. doi: 10.1158/1078-0432.CCR-05-0092. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Wobus M, Rangwala R, Sheyn I, Hennigan R, Coila B, Lower EE, Yassin RS, Sherman LS. CD44 associates with EGFR and erbB2 in metastasizing mammary carcinoma cells. Appl Immunohistochem Mol Morphol. 2002;10:34–39. doi: 10.1097/00129039-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y, Liu M, Yang B, Li B, Lu J. Role of siRNA silencing of MMP-2 gene on invasion and growth of laryngeal squamous cell carcinoma. Eur Arch Otorhinolaryngol. 2008;265:1385–1391. doi: 10.1007/s00405-008-0684-y. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Qiu J, Yang D, Lu J, Yan C, Zha X, Yin Y. MDM2 promotes invasion and metastasis in invasive ductal breast carcinoma by inducing matrix metalloproteinase-9. PLoS One. 2013;8:e78794. doi: 10.1371/journal.pone.0078794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao H, Zhang X, Zheng Y, Peng L, Hou J, Meng H. S100A9-induced release of interleukin (IL)-6 and IL-8 through toll-like receptor 4 (TLR4) in human periodontal ligament cells. Mol Immunol. 2015;67:223–232. doi: 10.1016/j.molimm.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Chen B, Miller AL, Rebelatto M, Brewah Y, Rowe DC, Clarke L, Czapiga M, Rosenthal K, Imamichi T, Chen Y, et al. S100A9 induced inflammatory responses are mediated by distinct damage associated molecular patterns (DAMP) receptors in vitro and in vivo. PLoS One. 2015;10:e0115828. doi: 10.1371/journal.pone.0115828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: An update. Biochim Biophys Acta. 2009;1793:993–1007. doi: 10.1016/j.bbamcr.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 27.Parkkila S, Pan PW, Ward A, Gibadulinova A, Oveckova I, Pastorekova S, Pastorek J, Martinez AR, Helin HO, Isola J. The calcium-binding protein S100P in normal and malignant human tissues. BMC Clin Pathol. 2008;8:2. doi: 10.1186/1472-6890-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang L, Lai YK, Zhang J, Wang H, Lin MC, He ML, Kung HF. Targeting S100P inhibits colon cancer growth and metastasis by Lentivirus-mediated RNA interference and proteomic analysis. Mol Med. 2011;17:709–716. doi: 10.2119/molmed.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dykxhoorn DM, Lieberman J. Knocking down disease with siRNAs. Cell. 2006;126:231–235. doi: 10.1016/j.cell.2006.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sy SM, Wong N, Lee TW, Tse G, Mok TS, Fan B, Pang E, Johnson PJ, Yim A. Distinct patterns of genetic alterations in adenocarcinoma and squamous cell carcinoma of the lung. Eur J Cancer. 2004;40:1082–1094. doi: 10.1016/j.ejca.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 31.Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol. 2012;24:277–283. doi: 10.1016/j.ceb.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ang KK, Berkey BA, Tu X, Zhang HZ, Katz R, Hammond EH, Fu KK, Milas L. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62:7350–7356. [PubMed] [Google Scholar]
- 33.Wobus M, Rangwala R, Sheyn I, Hennigan R, Coila B, Lower EE, Yassin RS, Sherman LS. CD44 associates with EGFR and erbB2 in metastasizing mammary carcinoma cells. Appl Immunohistochem Mol Morphol. 2002;10:34–39. doi: 10.1097/00129039-200203000-00006. [DOI] [PubMed] [Google Scholar]
- 34.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 35.Kivisaari AK, Kallajoki M, Ala-aho R, McGrath JA, Bauer JW, Königová R, Medvecz M, Beckert W, Grénman R, Kähäri VM. Matrix metalloproteinase-7 activates heparin-binding epidermal growth factor-like growth factor in cutaneous squamous cell carcinoma. Br J Dermatol. 2010;163:726–735. doi: 10.1111/j.1365-2133.2010.09924.x. [DOI] [PubMed] [Google Scholar]
- 36.Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, Elliston K, Stern D, Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- 37.Barclay AN. Membrane proteins with immunoglobulin-like domains-a master superfamily of interaction molecules. Semin Immunol. 2003;15:215–223. doi: 10.1016/S1044-5323(03)00047-2. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt AM, Yan SD, Yan SF, Stern DM. The biology of the receptor for advanced glycation end products and its ligands. Biochim Biophys Acta. 2000;1498:99–111. doi: 10.1016/S0167-4889(00)00087-2. [DOI] [PubMed] [Google Scholar]
- 39.Arumugam T, Simeone DM, Schmidt AM, Logsdon CD. S100P stimulates cell proliferation and survival via receptor for activated glycation end products (RAGE) J Biol Chem. 2004;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]