Abstract
A long non-coding RNA named HOXA transcript at the distal tip (HOTTIP) has been reported to be significantly increased in several cancers, including hepatocellular cancer, pancreatic cancer and lung cancer. However, the clinical value of HOTTIP expression in gastric cancer remains unknown. The present study aimed to investigate HOTTIP expression levels in gastric cancer and to elucidate its clinical significance. Reverse transcription polymerase chain reaction was used to assess the expression level of HOTTIP in gastric cancer cell lines and tissues. In a cohort of 94 patients with gastric cancer, HOTTIP expression was significantly lower in cancer tissues compared with the normal adjacent tissues. In addition, receiver operating characteristic (ROC) curve analysis was performed to evaluate the diagnostic value of HOTTIP in gastric cancer, and the area under the ROC curve was 0.767. In conclusion, the results of the present study indicated that HOTTIP may be a predictive biomarker for gastric cancer.
Keywords: biomarker, gastric cancer, HOXA transcript at the distal tip, long non-coding RNA
Introduction
Gastric cancer is one of the most frequent causes of cancer-associated mortality worldwide, with ~951,600 new cases and 723,100 mortalities occurring in 2012 (1). Although developments in early diagnosis, surgery, adjuvant chemotherapy and targeted therapies have improved, the prognosis of patients with gastric cancer and their long-term survival remains unsatisfactory (2). Numerous patients are diagnosed at a late stage and have a poor prognosis. Therefore, it is important and urgent to distinguish novel biomarkers in patients with gastric cancer for early diagnosis and to search for potential therapeutic targets.
Long non-coding RNAs (lncRNAs) are defined as transcripts containing >200 nucleotides without the capacity for coding proteins (3,4). A number of lncRNAs have been demonstrated to serve important functions in a wide range of diseases, including neurodegenerative diseases, chronic obstructive pulmonary disease and cancers (5,6). LncRNAs may function as oncogenes or tumor suppressor genes, and may be involved in the development or progression of cancers. Thus, lncRNAs may be used as cancer biomarkers for early diagnosis, as potential therapeutic targets and to predict cancer prognosis. Among these, homeobox (HOX)-associated lncRNAs are biologically important. For instance, HOX transcript antisense RNA (HOTAIR) and HOXA transcript at the distal tip (HOTTIP) are the most frequently studied in this area. HOTAIR is a lncRNA of 2,158 nucleotides in length, which is expressed in the HOXC locus of chromosome 12 (7). It has been reported to be a pro-oncogenic factor and a negative prognostic factor in several types of cancer, including breast, pancreatic, gastric, colorectal and bladder cancer (8–12).
Another lncRNA, HOTTIP, is an antisense non-coding transcript located at the 5′ end of the HOXA gene cluster. It has been reported to directly bind the adaptor protein WD repeat-containing protein 5 (WDR5) and to target WDR5/mixed lineage leukemia complexes, driving histone H3 lysine 4 trimethylation and gene transcription of distal HOXA genes (13). Several previous studies have reported that, compared with normal adjacent tissues (NATs), HOTTIP expression was significantly increased in skin, hepatocellular, pancreatic, lung and tongue squamous cell cancer tissues (14–20). HOTTIP may be involved in the progression of these cancers. However, the association between HOTTIP and gastric cancer remains unknown.
In the present study, the expression of HOTTIP was explored in gastric cancer tissues, and the associations between HOTTIP expression and clinicopathological characteristics were investigated.
Materials and methods
Patients and tissue samples
A total of 94 fresh gastric cancer tissues and matched NATs were obtained from patients who underwent radical resection for gastric cancer at the First Hospital of China Medical University (Shenyang, China) between May 2009 and July 2010. The matched NATs were obtained from tissues that were at least 5 cm from the edge of the cancer tissue. All tissues were frozen immediately in liquid nitrogen following resection and stored at −80°C prior to use. The tumor histological grade was classified according to the 7th edition of the Tumor-Node-Metastasis staging system (21).
The present study was approved by the Research Ethics Committee of China Medical University (Shenyang, China). Written informed consent was obtained from all patients.
Cell culture
The human gastric cancer cell lines MGC-803, BGC-823, SGC-7901 and HGC-27 were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The human gastric cancer AGS cell line was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). MGC-803, BGC-823, SGC-7901 and HGC-27 were cultured in RPMI-1640 medium (Biological Industries, Kibbutz Beit-Haemek, Israel). AGS was cultured in F-12K Medium (ATCC). Media were supplemented with 10% fetal bovine serum (Clark Bioscience, Claymont, DE, USA). Cell lines were cultured in an incubator at 37°C in a humidified atmosphere containing 5% CO2.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue samples and five cell lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The concentration and purity of total RNA was determined by using a Nano-Photometer UV/Vis spectrophotometer (Implen GmbH, München, Germany). A value of A260/A280>1.9 indicated good purity. Reverse transcription was performed using the PrimeScript RT reagent kit with gDNA eraser (Takara Biotechnology Co., Ltd., Dalian, China). qPCR analyses were performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) on a light cycler 480 II real-time PCR system (Roche Diagnostics, Basel, Switzerland). Each 25-µl PCR reaction mixture contained 0.3 µl forward and 0.3 µl reverse primers, 12.5 µl SYBR-Green mix, 2 µl gene cDNA and 9.9 µl RNase-free water. The reaction was amplified in 45 cycles of 95°C for 5 sec, 60°C for 30 sec and 72°C for 30 sec. Specific primers for HOTTIP were based on the first and fifth splice variant. The sequences of the primers used were as follows: HOTTIP forward, 5′-CGTAGAGACACAGGCAGCAG-3′ and reverse, 5′-CAGCCGAACAGAGTCAGAGG-3′; GAPDH, forward 5′-CGGATTTGGTCGTATTGGG-3′ and reverse, 5′-CTGGAAGATGGTGATGGGATT-3′. The RT-qPCR reactions were performed in triplicate. All reagents were used according to the manufacturer's protocol. The expression level of HOTTIP in gastric cancer tissues compared with NATs was calculated using the 2−ΔΔCq method (22). If the 2−ΔΔCq value was <1, there was low expression in cancer tissues and cancer cell lines compared with NATs.
Statistical analysis
Statistical analysis was performed using SPSS software version 17.0 (SPSS, Inc., Chicago, IL, USA). The associations between HOTTIP expression and clinicopathological characteristics were tested using non-parametric tests: The Mann-Whitney U test for 2 groups and the Kruskal-Wallis test for ≥3 groups. Survival rates, including overall survival and disease-free survival were calculated using the Kaplan-Meier method with the log-rank test applied for comparison. A receiver operating characteristic (ROC) curve was used to evaluate the diagnostic value of HOTTIP expression levels. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of HOTTIP in gastric cancer
Using RT-qPCR, significantly decreased HOTTIP expression levels were observed in MGC-803 cells, BGC-823 cells, SGC-7901 cells, HGC-27 and AGS cells compared with three NATs randomly selected from the patients (P<0.001; Fig. 1).
Figure 1.
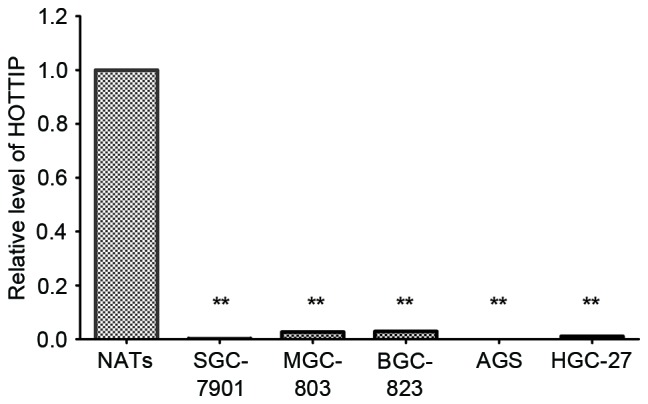
Expression of HOTTIP in five gastric cancer cell lines. Quantification of HOTTIP was detected by reverse transcription-quantitative polymerase chain reaction. Data are presented in gastric cancer cell lines relative to three NATs randomly selected from 94 patients as a control. Data were expressed as the mean ± standard deviation from at least three separate experiments. **P<0.01 vs. NATs. HOTTIP, HOXA transcript at the distal tip; NATs, normal adjacent tissues.
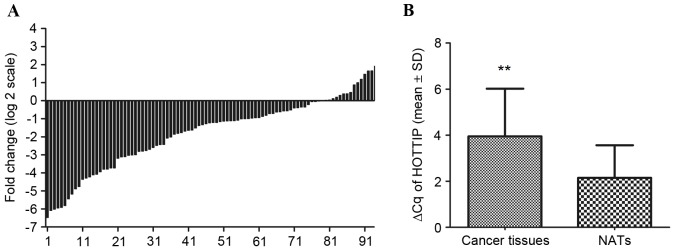
In addition, HOTTIP was detected in all 94 gastric cancer tissues and their paired NATs. Among the 94 patients with gastric cancer, 83% (78/94) of cases indicated that the expression of HOTTIP was decreased in gastric cancer tissues compared with their paired NATs (Fig. 2A). Furthermore, HOTTIP expression was significantly decreased in cancer tissues compared with NATs (P<0.001; Fig. 2B).
Figure 2.
Expression of HOTTIP in 94 patients with gastric cancer. (A) Data were presented as log2 of fold-change of gastric cancer tissues relative to NATs. Each case was analyzed in triplicate and repeated three times. (B) ΔCq was used to compare the relative expression of HOTTIP in cancer tissues and NATs. The results indicated that the value of ΔCq was significantly higher in gastric cancer tissues compared with NATs. Data were expressed as the mean ± standard deviation from at least three separate experiments. Larger ΔCq values indicated lower expression. **P<0.01 vs. NATs. HOTTIP, HOXA transcript at the distal tip; NATs, normal adjacent tissues.
Associations between HOTTIP expression and clinicopathological characteristics
The present study also investigated the associations between HOTTIP expression and clinicopathological parameters using non-parametric tests. No statistically significant association was observed between HOTTIP expression and any clinicopathological characteristic (Table I).
Table I.
Associations between the expression of HOTTIP and clinicopathological characteristics in 94 patients with gastric cancer.
| Clinicopathological characteristic | n | HOTTIP expressiona | P-value |
|---|---|---|---|
| Gender | 0.892 | ||
| Male | 68 | 0.389 (0.124–0.782) | |
| Female | 26 | 0.463 (0.096–0.675) | |
| Age, years | 0.218 | ||
| ≥63 | 48 | 0.301 (0.117–0.654) | |
| <63 | 46 | 0.465 (0.121–0.967) | |
| Tumor size, cm | 0.145 | ||
| ≥5.5 | 56 | 0.301 (0.084–0.744) | |
| <5.5 | 38 | 0.498 (0.175–0.760) | |
| Macroscopic typeb | 0.304 | ||
| Early stage | 6 | 0.466 (0.029–0.598) | |
| Borrmann I–II | 7 | 0.520 (0.463–1.234) | |
| Borrmann III–IV | 81 | 0.382 (0.119–0.725) | |
| Differentiation | 0.397 | ||
| Good | 3 | 0.292 (0.034–0.697) | |
| Moderate | 40 | 0.280 (0.062–0.774) | |
| Poor | 51 | 0.460 (0.154–0.760) | |
| Lauren typec | 0.228 | ||
| Intestinal type | 32 | 0.256 (0.054–0.774) | |
| Diffuse type | 62 | 0.450 (0.152–0.695) | |
| pT stage | 0.578 | ||
| T1+T2 | 14 | 0.506 (0.315–0.977) | |
| T3 | 16 | 0.335 (0.139–0.926) | |
| T4 | 64 | 0.389 (0.111–0.739) | |
| pN stage | 0.512 | ||
| N0 | 20 | 0.390 (0.083–0.516) | |
| N1 | 17 | 0.609 (0.195–1.008) | |
| N2 | 20 | 0.270 (0.128–0.706) | |
| N3 | 37 | 0.445 (0.117–0.972) | |
| pTNM stage | 0.431 | ||
| I | 8 | 0.447 (0.019–0.508) | |
| II | 19 | 0.499 (0.185–0.964) | |
| III | 67 | 0.396 (0.116–0.760) |
Median relative expression (25–75th percentile)
see reference 38
see reference 39. n, number of patients; p, pathological; HOTTIP, HOXA transcript at the distal tip; T, tumor; N, node; M, metastasis.
Association between HOTTIP expression and patient survival time
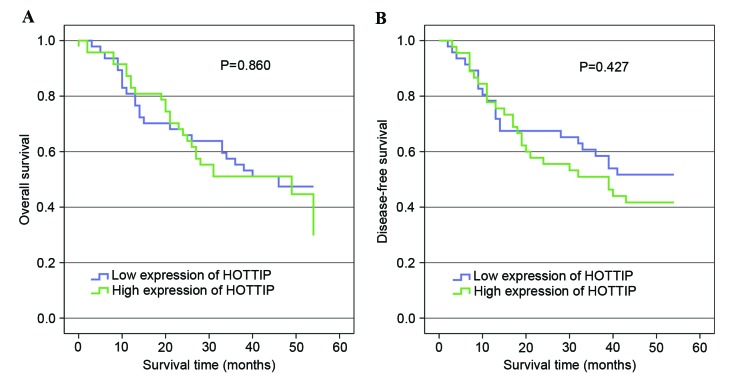
To investigate the association between HOTTIP expression and the survival time of patients with gastric cancer, Kaplan-Meier analysis was performed. The median relative expression levels of HOTTIP were used to divide patients into high and low expression groups. Kaplan-Meier analyses indicated there was no significant prognostic difference between patients with high and low HOTTIP expression (Fig. 3).
Figure 3.
HOTTIP expression and survival. (A) Kaplan-Meier analysis of overall survival based on HOTTIP expression in all 94 patients with gastric cancer (P=0.860). (B) Kaplan-Meier analysis of disease-free survival based on HOTTIP expression in all 94 patients with gastric cancer (P=0.427). HOTTIP, HOXA transcript at the distal tip.
The diagnostic value of using HOTTIP as a biomarker
To evaluate the diagnostic value of HOTTIP for distinguishing gastric cancer tissue from normal tissue, a ROC curve was constructed. The area under the ROC curve (AUC) was 0.767 (P<0.001; Fig. 4), which indicated that HOTTIP is a potential biomarker for gastric cancer.
Figure 4.
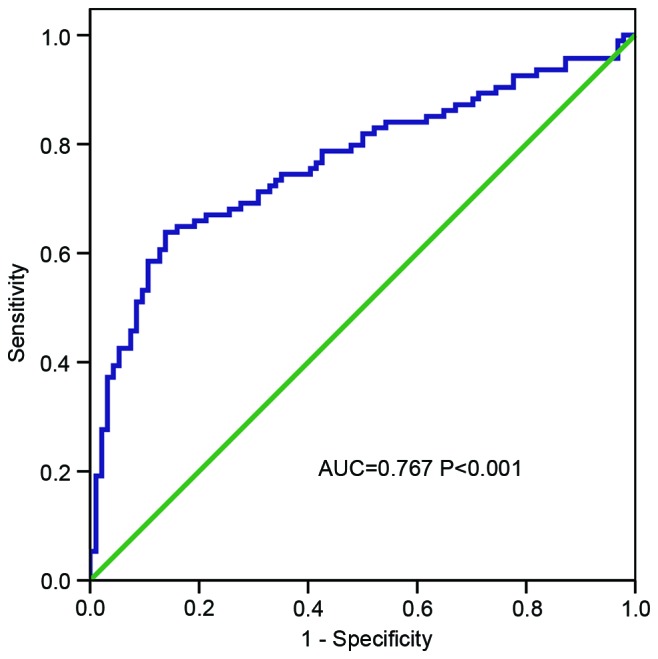
ROC curve based on HOXA transcript at the distal tip expression in cancer tissues and normal adjacent tissues. The AUC was 0.767 (P<0.001). ROC, Receiver operating characteristic; AUC, area under ROC curve.
Discussion
The prognosis of gastric cancer remains poor as the majority of patients are diagnosed at a late stage, when treatments are less effective (2,23). Thus, cancer screening and early diagnosis serve an important function in the survival of patients with gastric cancer. The identification of novel and specific biomarkers is of great clinical value for the diagnosis and treatment of gastric cancer. Multiple previous studies have reported that lncRNAs are involved in cancer pathogenesis and may provide novel insight into the biology of this disease (5,6), where they may act as oncogenes or tumor suppressors. A number of previous studies have reported that HOTAIR may possess pro-oncogenic functions and that it is involved in multiple cancers (8–12). Similarly, a number of studies have demonstrated that HOTTIP may serve as a critical regulator in certain cancers. For example, Quagliata et al (15) reported that HOTTIP and HOXA13 were highly expressed in patients with hepatocellular carcinoma, where they were associated with metastasis and poor survival. Li et al (17) reported that HOTTIP was upregulated in pancreatic cancer and may promote cancer cell proliferation, invasion and chemoresistance by regulating HOXA13. However, another previous study reported that in pancreatic cancer cells, HOTTIP did not regulate HOXA13 but was involved in the regulation of certain HOX genes, including HOXA1, HOXA9, HOXA10, HOXA11 and HOXB2 (16). Deng et al (19) reported that HOTTIP promoted tumor growth and inhibited cell apoptosis in lung cancer. In conclusion, dysregulated expression of HOTTIP has been associated with a wide variety of biological characteristics of tumors, and exists in multiple cancers.
To the best of our knowledge, this is the first report to investigate the value of HOTTIP in gastric cancer. The present results indicated that HOTTIP expression was significantly downregulated in gastric cancer tissues compared with NATs among 94 patients (P<0.001). Furthermore, a ROC curve was constructed to evaluate the diagnostic value of HOTTIP in gastric cancer. The results revealed that the AUC was 0.767, suggesting that HOTTIP had potential diagnostic value in gastric cancer. Previously, multiple studies reported that in different types of cancer there was abnormal expression of lncRNA, indicating that the same lncRNA may serve different functions in different types of cancer. For instance, a lncRNA named SPRY4 intronic transcript 1 (SPRY4-IT1) was reported to be overexpressed in melanoma, renal cell carcinoma, esophageal squamous cell carcinoma, breast cancer, bladder cancer and glioma, and associated with poor prognosis and promotion of tumor growth (24–29). Certain studies also demonstrated that SPRY4-IT1 expression was decreased in non-small-cell lung cancer and gastric cancer, and acted with significant antitumor function in these two types of cancer (30,31). Similarly, HOTTIP may exhibit a tissue-specific expression pattern and serve a different function in different types of cancer, as with SPRY4-IT.
A number of studies have reported that lncRNAs may act on their neighboring protein-coding genes in a cis-manner (32,33). HOTTIP is located the 5′ end of the HOXA gene cluster, and Wang et al (13) demonstrated that chromosomal looping may make HOTTIP in close proximity to gene targets, thus, HOTTIP may be involved in the regulation of its neighboring HOXA genes. It was reported that HOTTIP may regulate the expression of HOX genes, including HOXA1, HOXA9, HOXA10, HOXA11 and HOXA13, in liver cancer and pancreatic cancer (15–17). In addition, numerous studies have reported that aberrant expression of HOXA genes was associated with a number of biological characteristics of gastric cancer (34–37). HOTTIP may be critical in regulating HOXA genes in gastric cancer, but the underlying mechanism remains unknown and requires additional study.
In summary, the results of the present study indicated that lncRNA HOTTIP expression was decreased in patients with gastric cancer, and that HOTTIP may be a predictive biomarker in gastric cancer. However, the underlying molecular mechanisms through which HOTTIP is involved in gastric cancer require further study.
Acknowledgements
The authors would like to thank the Department of Surgical Oncology and General Surgery, First Hospital of China Medical University (Shenyang, China) for providing the human gastric tissue samples. The authors would also like to thank the College of China Medical University (Shenyang, China) for technical assistance in experiments. The present study was supported by the National Science Foundation of China (grant nos. 81201888, 81372549 and 81172370), the Natural Science Foundation of Liaoning Province (grant no. 2014029201) and the Key Technologies Research and Development Program of Liaoning Province (grant no. 2012225008).
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Shen L, Shan YS, Hu HM, Price TJ, Sirohi B, Yeh KH, Yang YH, Sano T, Yang HK, Zhang X, et al. Management of gastric cancer in Asia: Resource-stratified guidelines. Lancet Oncol. 2013;14:e535–e547. doi: 10.1016/S1470-2045(13)70436-4. [DOI] [PubMed] [Google Scholar]
- 3.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 4.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 6.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao W, Dong S, Duan B, Chen P, Shi L, Gao H, Qi H. HOTAIR is a predictive and prognostic biomarker for patients with advanced gastric adenocarcinoma receiving fluorouracil and platinum combination chemotherapy. Am J Transl Res. 2015;7:1295–1302. [PMC free article] [PubMed] [Google Scholar]
- 9.Xue Y, Gu D, Ma G, Zhu L, Hua Q, Chu H, Tong N, Chen J, Zhang Z, Wang M. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2015;30:303–310. doi: 10.1093/mutage/geu076. [DOI] [PubMed] [Google Scholar]
- 10.Sun X, Du P, Yuan W, Du Z, Yu M, Yu X, Hu T. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 2015;6:e1907. doi: 10.1038/cddis.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang YJ, Bikle DD. LncRNA profiling reveals new mechanism for VDR protection against skin cancer formation. J Steroid Biochem Mol Biol. 2014;144:87–90. doi: 10.1016/j.jsbmb.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A, Kovac M, Moretti F, Makowska Z, Boldanova T, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget. 2015;6:10840–10852. doi: 10.18632/oncotarget.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H, Wang Y, Zeng J, Song Y, Gao W, et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J Transl Med. 2015;13:84. doi: 10.1186/s12967-015-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsang FH, Au SL, Wei L, Fan DN, Lee JM, Wong CC, Ng IO, Wong CM. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 2015;35:1597–1606. doi: 10.1111/liv.12746. [DOI] [PubMed] [Google Scholar]
- 19.Deng HP, Chen L, Fan T, Zhang B, Xu Y, Geng Q. Long non-coding RNA HOTTIP promotes tumor growth and inhibits cell apoptosis in lung cancer. Cell Mol Biol (Noisy-le-grand) 2015;61:34–40. [PubMed] [Google Scholar]
- 20.Zhang H, Zhao L, Wang YX, Xi M, Liu SL, Luo LL. Long non-coding RNA HOTTIP is correlated with progression and prognosis in tongue squamous cell carcinoma. Tumour Biol. 2015;36:8805–8809. doi: 10.1007/s13277-015-3645-2. [DOI] [PubMed] [Google Scholar]
- 21.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. American Joint Committee on Cancer (AJCC) Cancer Staging Handbook. 7th. Springer-Verlag; New York: 2010. [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477–490. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 25.Xie HW, Wu QQ, Zhu B, Chen FJ, Ji L, Li SQ, Wang CM, Tong YS, Tuo L, Wu M, et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol. 2014;35:7743–7754. doi: 10.1007/s13277-014-2013-y. [DOI] [PubMed] [Google Scholar]
- 26.Zhang HM, Yang FQ, Yan Y, Che JP, Zheng JH. High expression of long non-coding RNA SPRY4-IT1 predicts poor prognosis of clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2014;7:5801–5809. [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H, Lv Z, Guo E. Knockdown of long noncoding RNA SPRY4-IT1 suppresses glioma cell proliferation, metastasis and epithelial-mesenchymal transition. Int J Clin Exp Pathol. 2015;8:9140–9146. [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Y, Li J, Liu Y, Ding J, Fan Y, Tian Y, Wang L, Lian Y, Wang K, Shu Y. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer. 2015;14:51. doi: 10.1186/s12943-015-0318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao XL, Zhao ZH, Xu WC, Hou JQ, Du XY. Increased expression of SPRY4-IT1 predicts poor prognosis and promotes tumor growth and metastasis in bladder cancer. Int J Clin Exp Pathol. 2015;8:1954–1960. [PMC free article] [PubMed] [Google Scholar]
- 30.Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, Yang JS, Xu TP, Liu YW, Zou YF, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298. doi: 10.1038/cddis.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie M, Nie FQ, Sun M, Xia R, Liu YW, Zhou P, De W, Liu XH. Decreased long noncoding RNA SPRY4-IT1 contributing to gastric cancer cell metastasis partly via affecting epithelial-mesenchymal transition. J Transl Med. 2015;13:250. doi: 10.1186/s12967-015-0595-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelsey AD, Yang C, Leung D, Minks J, Dixon-McDougall T, Baldry SE, Bogutz AB, Lefebvre L, Brown CJ. Impact of flanking chromosomal sequences on localization and silencing by the human non-coding RNA XIST. Genome Biol. 2015;16:208. doi: 10.1186/s13059-015-0774-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guil S, Esteller M. Cis-acting noncoding RNAs: Friends and foes. Nat Struct Mol Biol. 2012;19:1068–1075. doi: 10.1038/nsmb.2428. [DOI] [PubMed] [Google Scholar]
- 34.Bai Y, Fang N, Gu T, Kang Y, Wu J, Yang D, Zhang H, Suo Z, Ji S. HOXA11 gene is hypermethylation and aberrant expression in gastric cancer. Cancer Cell Int. 2014;14:79. doi: 10.1186/s12935-014-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han Y, Tu WW, Wen YG, Li DP, Qiu GQ, Tang HM, Peng ZH, Zhou CZ. Identification and validation that up-expression of HOXA13 is a novel independent prognostic marker of a worse outcome in gastric cancer based on immunohistochemistry. Med Oncol. 2013;30:564. doi: 10.1007/s12032-013-0564-1. [DOI] [PubMed] [Google Scholar]
- 36.Degl' Innocenti D Rossi, Castiglione F, Buccoliero AM, Bechi P, Taddei GL, Freschi G, Taddei A. Quantitative expression of the homeobox and integrin genes in human gastric carcinoma. Int J Mol Med. 2007;20:621–629. [PubMed] [Google Scholar]
- 37.Sentani K, Oue N, Naito Y, Sakamoto N, Anami K, Oo HZ, Uraoka N, Aoyagi K, Sasaki H, Yasui W. Upregulation of HOXA10 in gastric cancer with the intestinal mucin phenotype: Reduction during tumor progression and favorable prognosis. Carcinogenesis. 2012;33:1081–1088. doi: 10.1093/carcin/bgs121. [DOI] [PubMed] [Google Scholar]
- 38.Borrmann R. Geshwulste des magens und duodenums. In: Henke F, Lubarsch O, editors. Handbuch der speziellen pathologischen anatomie und histologie. Springer-Verlag; Berlin: 1926. p. 865. [Google Scholar]
- 39.Lauren P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]