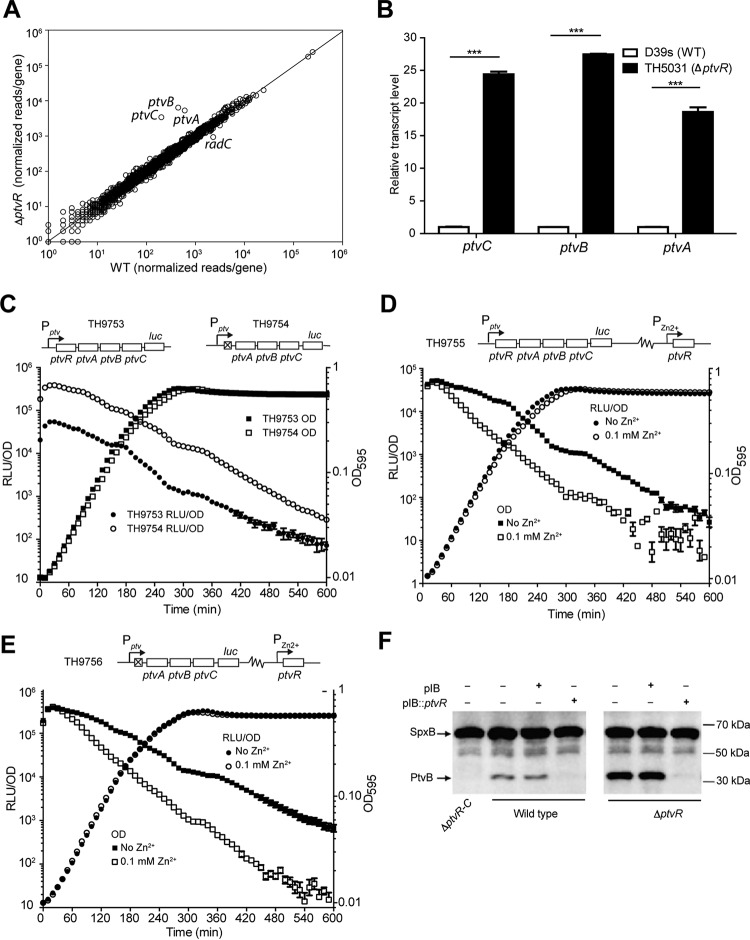
FIG 2.
Transcriptional repression of the ptv operon by PtvR. (A) Comparison of the transcriptomes of strain D39s and the isogenic ptvR-deficient (ΔptvR) strain by RNA-Seq. Reads per gene were normalized to reads per kilobase per million reads. The line represents y = x. (B) Transcription derepression of the ptv operon in the ΔptvR strain. The transcripts of ptvA, ptvB, and ptvC in the ΔptvR mutant strain were measured by qRT-PCR and normalized to the corresponding value of each gene in D39s (WT), as for Fig. 1B. ***, P < 0.001. (C) Transcriptional repression of the ptv locus by PtvR as assessed by fusing the luc reporter gene to the 3′ end of ptvC in wild-type (TH9753, Pptv_luc) or ΔptvR (TH9754, Pptv_luc) cells. The cell density (OD595) and luciferase activity (RLU/OD) were measured every 10 min for 10 h. (D and E) Additional repression of the ptv locus by trans overexpression of ptvR. The OD595 and RLU/OD values in the presence (D) or absence (E) of the endogenous ptvR were measured with or without Zn2+, as for panel C. (F) Detection of transcription repression by PtvR at the protein level. PtvB was detected by immunoblotting in D39s (wild type), ΔptvR-C, and ΔptvR in the presence (+) or absence (-) of the empty pIB166 (pIB) and the ptvR complementation construct (pIB::ptvR). SpxB was similarly detected as a loading control. The sizes of the protein standards are indicated.