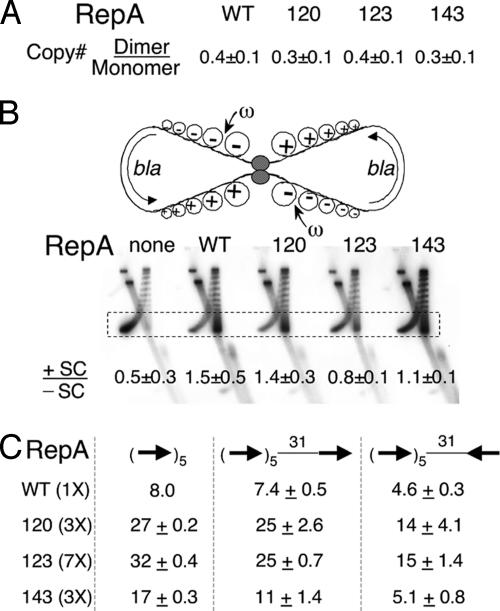
Fig. 2.
Handcuffing efficiency of RepA mutants in vivo. (A) Copy number ratio of isogenic dimeric and monomeric forms of miniP1 plasmids when the RepA protein is WT or mutant 120, 123, or 143. (B) A topological assay for interactions between two arrays of five consensus iterons of a homodimeric plasmid derived from pBR322. The semicircular arrows show the direction of transcription of the plasmid bla gene. Transcription-generated positive (+) and negative (–) supercoils (SC) are shown. The small arrow indicates that the ω protein targets the –SC. The topoisomers are separated on a 2D gel. In the first dimension, +SC and –SC run together. In the second dimension, when chloroquine was present, +SC run ahead of –SC. The boxed region of the distribution, where the SC are well separated, was used to calculate the ratio of +SC and –SC as described in ref. 25. (C) MiniP1 copy number with an extra iteron placed 31 bp downstream of the five origin iterons in two orientations. The WT and mutant RepAs were supplied from constitutive sources in different amounts to achieve similar levels of iteron binding. Copy number of miniP1 without any extra iteron in the presence of 1× WT RepA was taken as 8.0 (30).