Figure 1.
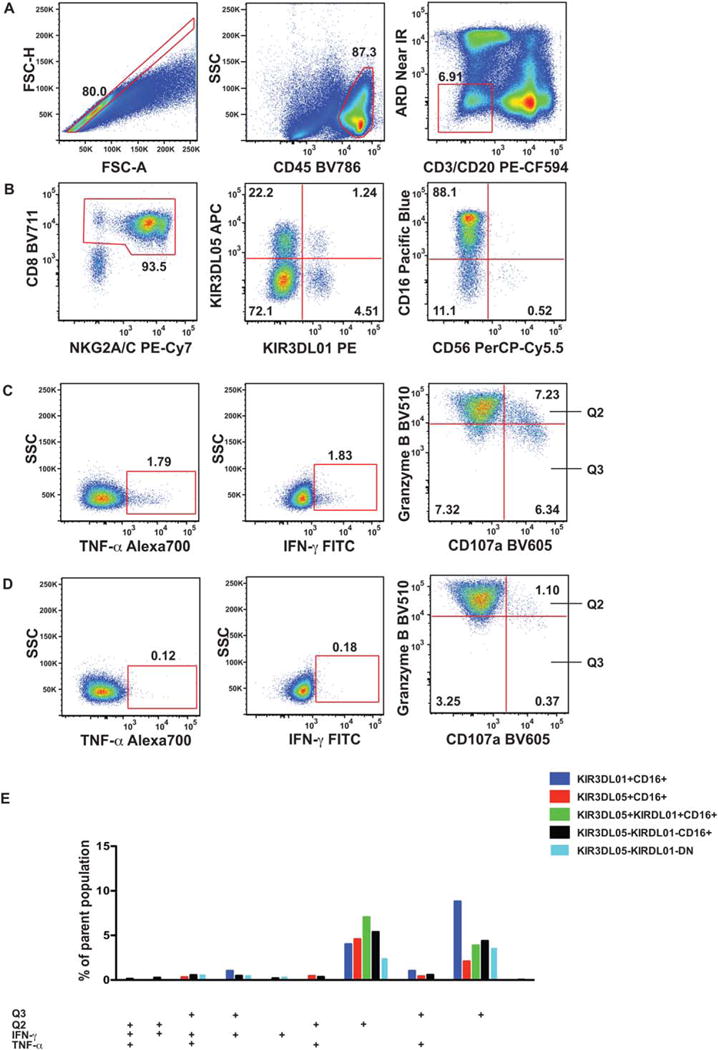
Gating strategy for OMIP-035. Ficoll-purified PBMC of an SIVmac239-infected rhesus macaque were stimulated with 721.221 cells for 18 h, then stained with the NK cell function panel as detailed in the Supporting Information. (A) With the initial sequential gating we excluded cell aggregates (single cell gate defined by forward scatter area and height), dead cells (Amine reactive dye−), T cells (CD3−), B cells (CD20−), and human 721.221 cells (CD45+) gate. CD8+ and NKG2A/C+ antigens were used to define the NK cell population. (B) Within the NK cell population, four subsets were identified using the KIR3DL01 and KIR3DL05 receptor expression pattern. All four subsets were further divided into CD16+, CD56+, and CD16-CD56− (DN) populations. (The third panel shows the KIR3D double negative population.) (C) Next, we defined the IFN-γ+, TNF-α+, CD107a+ Granzyme Bhigh (Q2), and CD107a+ Granzyme Blow (Q3) subsets using (D) the matching non-stimulated sample within the CD16+, and CD16-CD56− NK cell populations. (Panels in the C and D series shows the CD16+ subset within the KIR3D double negative population.) (E) Functional subpopulations, as defined by the combined expression pattern of IFN-γ+, TNF-α+, CD107a + Granzyme Bhigh (Q2), and CD107a+ Granzyme Blow (Q3) subsets were quantified by Boolean gating and displayed as a bar graph. Only subsets above 0.6% of the parent population, containing >10 events are displayed. None of the CD56+ populations reached this cutoff level.
