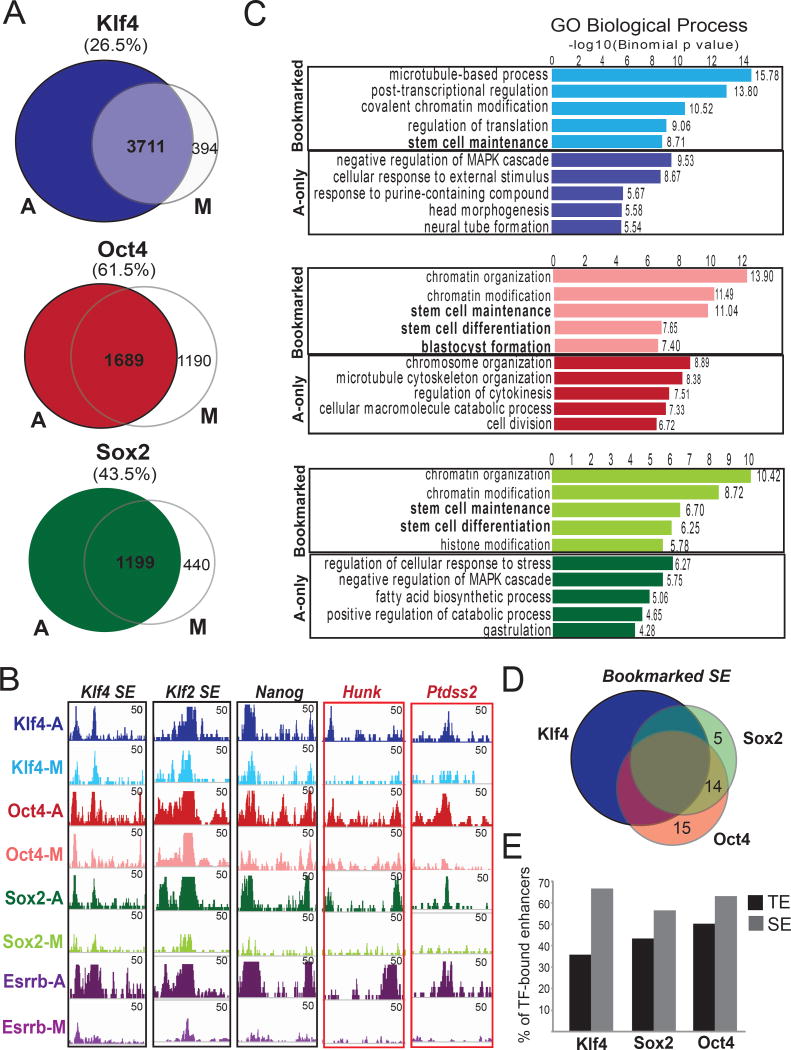
Figure 3. ChIP-seq assays in mitotic and asynchronous ESCs identify the bookmarked genomic targets of KLF4, OCT4 and SOX2 (KOS).
A. Venn diagrams showing numbers of unique or common KOS-binding sites in asynchronous (A) and mitotic (M) mESCs. ChIP-seq peaks consistently detected in at least two of three biological replicates were used for the analyses (see Supplemental Material). The percent of asynchronous peaks that were also detected in mitotic cells (bookmarked) are reported in parentheses. B. Examples of genomic regions that retained (black outline) or lost (red outline) KOS binding during mitosis. Note that the extent of KOS retention was similar or higher compared to the previously reported bookmarking factor ESRRB (published datasets from Festuccia et al., 2016). C. Top 5 gene ontology categories (GO) enriched in KLF4 (blue), OCT4 (red) and SOX2 (green) binding sites that were either common between A and M (bookmarked) or present only in A (A-only). GOs associated with stem cell identity (bold) were overrepresented in bookmarked peaks. D. Venn diagram depicting the number of ESC-specific super-enhancers (SE) (Whyte et al., 2013) that remained bookmarked by individual TFs and combinations, highlighting the high frequency of combinatorial bookmarking. E. Barplot showing the percentage of KLF4, SOX2 or OCT4-bound typical enhancers (TE) and SE that remained bookmarked during mitosis.