Figure 1.
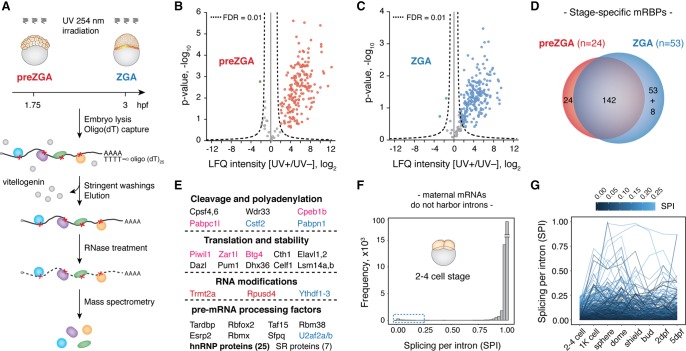
The zebrafish MZT mRNA-bound proteome is conserved, dynamic, and rich in pre-mRNA processing factors. (A) Schematic of the method for in vivo identification of mRBPs from early zebrafish embryos. Experiments were performed at two developmental time points: preZGA (32–64 cell stage) and ZGA (512–1k cell stage) in three biological replicates: (hpf) hours post-fertilization. (B,C) Volcano plots display the enrichment of detected proteins between UV+ and UV− samples (x-axis) and their significance (y-axis). Hyperbolic, dashed lines represent the applied statistical threshold of FDR = 0.01. Data points beyond this threshold refer to proteins enriched in UV+ (blue and red) and UV− (green) conditions. Nonenriched proteins are shown in gray. (D) Venn diagram displaying differences between preZGA (red) and ZGA (blue) mRBPomes. Note that eight proteins within the ZGA-specific group were also detected at the preZGA stage, yet below the enrichment threshold. (E) MZT mRBPome components grouped with respect to their major known functions. Names of the selected proteins are color-coded based on their embryo- and stage-specific detection: (black) both stages; (red) preZGA-specific; (blue) ZGA-specific; (pink) embryo-specific. (F) Histogram displaying splicing score per intron (SPI) of all expressed maternal mRNAs at the two- to four-cell stage shows that the majority (97.5%) of maternal transcripts are efficiently spliced. (G) Line plot tracking introns with low SPI values (0–0.25) from the two- to four-cell stage to 5 d post-fertilization (dpf) suggests the majority of these introns are never spliced out.