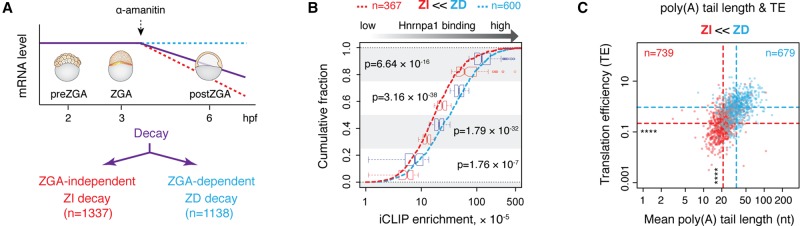
Figure 3.
Hnrnpa1 preferentially associates with ZGA-dependent decay genes. (A) Schematic representation of two distinct decaying gene groups. All decaying maternal mRNAs are shown by a purple line. Among those, ZGA-independent (ZI) decay genes (red dashed line) do not require zygotic transcription for their destabilization and continuously decay upon α-amanitin treatment. ZGA-dependent (ZD) decay genes (blue dashed line) require zygotic transcriptional activity for their clearance, thereby remaining stable in α-amanitin treated embryos. The total number of identified ZI and ZD decay genes is indicated. (B) Cumulative plot showing the fraction of ZI (red dashed line) and ZD (dashed blue line) decay genes (y-axis) and their respective Hnrnpa1 iCLIP enrichment (x-axis). Box plots in the background show the distribution of iCLIP enrichment per each quartile for ZI and ZD decay genes. Indicated P-values were derived from pairwise comparison of ZI (red) and ZD (blue) decay genes within each quartile (Wilcoxon rank test). (C) Scatterplot showing differences in poly(A) tail length and TE of ZI and ZD decay genes. Red and blue dashed lines refer to the median of poly(A) tail length and TE for ZI and ZD decay genes, respectively. The number of genes in each group is indicated: (****) P < 2.2 × 10−16 (Wilcoxon rank test).