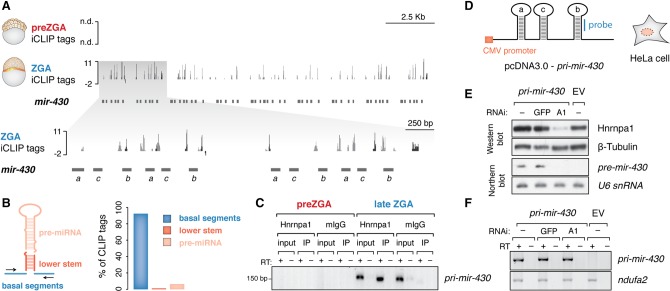
Figure 5.
Hnrnpa1 binds nuclear pri-mir-430 transcripts and regulates their processing at ZGA. (A) ZGA-specific Hnrnpa1 binding across the mir-430 locus. Significant iCLIP tags (FDR < 0.01) derived from unique and multimapped reads are shown in dark and light gray, respectively. The inset displays an enhanced view of iCLIP tags in the 5′ end of the mir-430 locus (gray shaded box). Annotations of individual mir-430 genes are indicated with horizontal dark gray bars (mir-430a, mir-430c, mir-430b): (n.d.) not detected; negative values in the tracks represent iCLIP tags mapped to the opposite (−) strand. (B) Bar plot shows relative abundance of Hnrnpa1 iCLIP tags derived from multimapped reads across different types of pri-mir-430 regions. Each bar color corresponds to the colored region in the schematic depiction of the pri-miRNA structure on the left. (C) RT-PCR on RNA extracted from in vivo crosslinked and immunopurified Hnrnpa1–RNA complexes shows specific association of the protein with pri-mir-430 transcripts only at the time of its expression. Note a lack of RNA detection at the preZGA stage. (D) Schematic of the cytomegalovirus (CMV) promoter-driven construct harboring three consecutive miR-430 hairpins (a, c, b) from the zebrafish miR-430 cluster utilized for transient HeLa cell transfection. Location of the DNA probe used for Northern blot analysis is shown in blue. (E) Western blot showing levels of Hnrnpa1 in cells transfected with shRNA-expressing constructs against Hnrnpa1 (A1), GFP (GFP), and empty RNAi vector (−). Cells were subsequently transfected with pri-mir-430 construct and empty vector (EV). Beta-tubulin served as a loading control. Northern blot showing a decrease in pre-mir-430 levels upon Hnrnpa1 depletion; U6 snRNA served as a loading control. Note that the human ortholog of mir-430 is not endogenously expressed (−/EV). (F) RT-PCR on total RNA from HeLa cells shows unchanged pri-mir-430 levels upon Hnrnpa1 depletion.