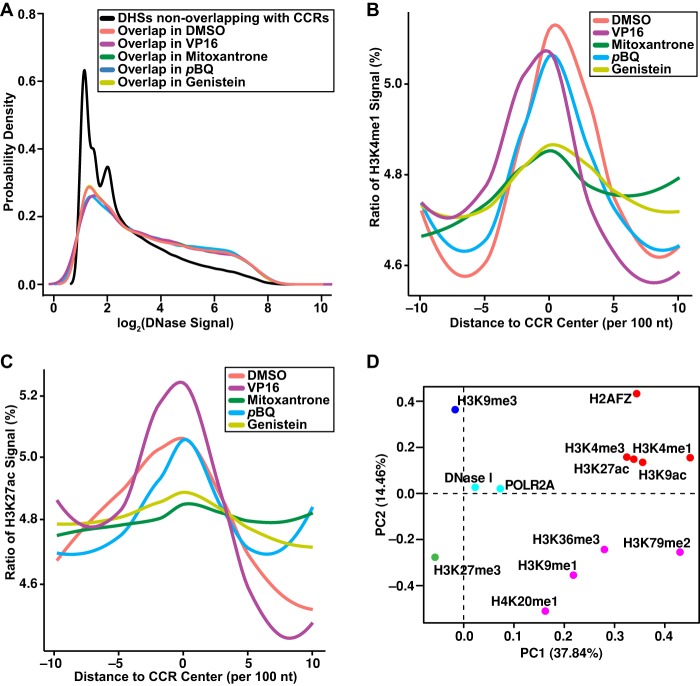
Figure 7.
Colocalization of genome-wide TOP2A CCRs with chromatin features. (A) Higher DNase I signal density in DHSs that overlap (colored lines) with CCRs compared with DHSs that do not (black line) overlap. P < 2.2 × 10−16; Kruskal–Wallis test. (B,C) Enriched H3K4me1 (B) and H3K27ac (C) signals at CCR centers (position 0 on x-axis) compared with 1-kb upstream and downstream flanking sequences in 100-nt sliding windows. P < 0.001 for DMSO, VP16, pBQ; P < 0.01 for mitoxantrone, genistein; Kruskal–Wallis test. (A–C) Amplified samples; same treatments merged where applicable. (D) Clustering of features found within CCRs by PCA and k-means algorithms. PC1 and PC2 account for 37.84% and 14.46% of the total variance, respectively. Dots represent PCA loadings for indicated features. Colors show five clusters of features found in CCRs. Along PC1, note the separation of histone marks of gene repression (dark blue, green) from marks known to positively affect gene expression (light blue, purple, red). Along PC2, note the separation between clusters of marks of actively expressed genes (light blue, purple) from cluster of gene activating elements including enhancer marks (red). Union set of CCRs from all amplified samples. (A–D) Analyses were performed on existing data for chromatin features (The ENCODE Project Consortium 2012) after liftOver (Hinrichs et al. 2006; http://genome.sph.umich.edu/wiki/LiftOver) conversion to GRCh38/hg38. See also Supplemental Tables S1, S4.