Figure 4.
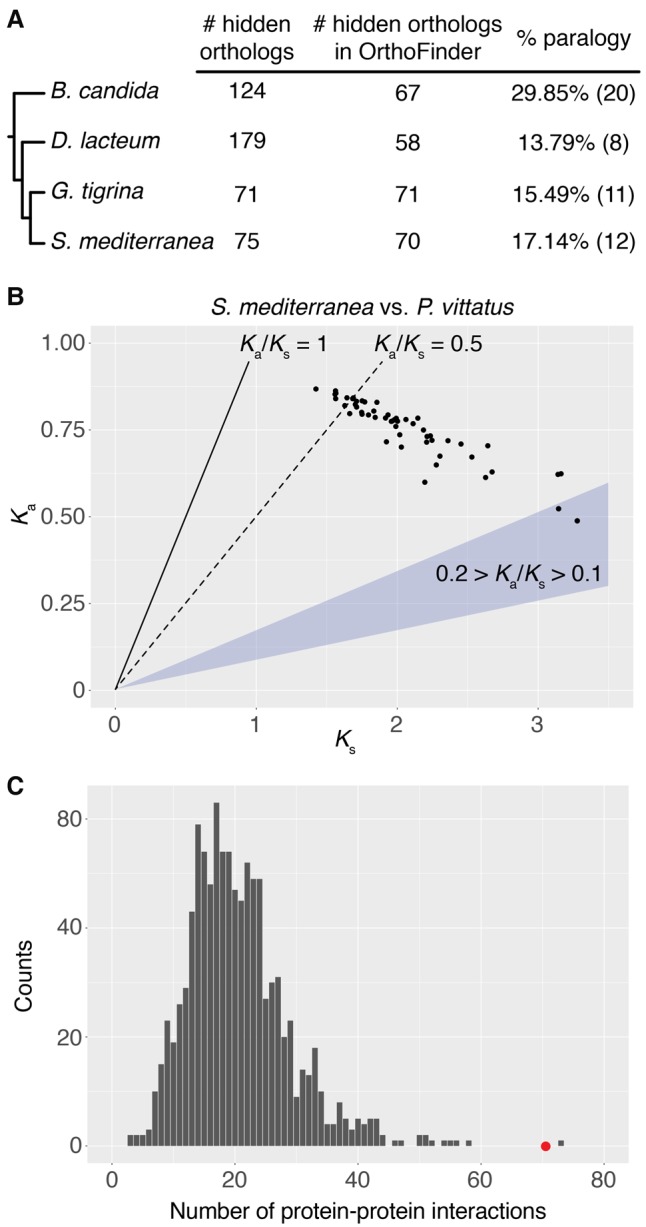
Level of paralogy and Ka/Ks values in triclad hidden orthologs. (A) Percentage of hidden orthologs identified by Leapfrog that are present in OrthoFinder and share an orthogroup with other sequences of the same species. We deem these cases as probable fast evolving paralogs (hidden paralogy). (B) Ka and Ks values of 53 one-to-one hidden orthologs of S. mediterranea compared with their respective homologs in the “bridge” species P. vittatus. Although in almost half of these hidden orthologs the Ks value suggested saturation (Ks > 2), for most of the rest the Ka/Ks value was above or around 0.5 (dotted line), which can be a sign of weak positive selection or relaxed constraint. (C) Number of predicted protein–protein interactions in S. mediterranea hidden orthologs (red dot) compared with a distribution of interactions observed in 1000 random samples of similar size (gray bars). Hidden orthologs show a significantly higher number of interactions, suggesting that complementary mutations between protein partners might drive hidden orthology in flatworms.
