Figure 2. Overview of the major microfluidic device types: valves, droplets and nanowells.
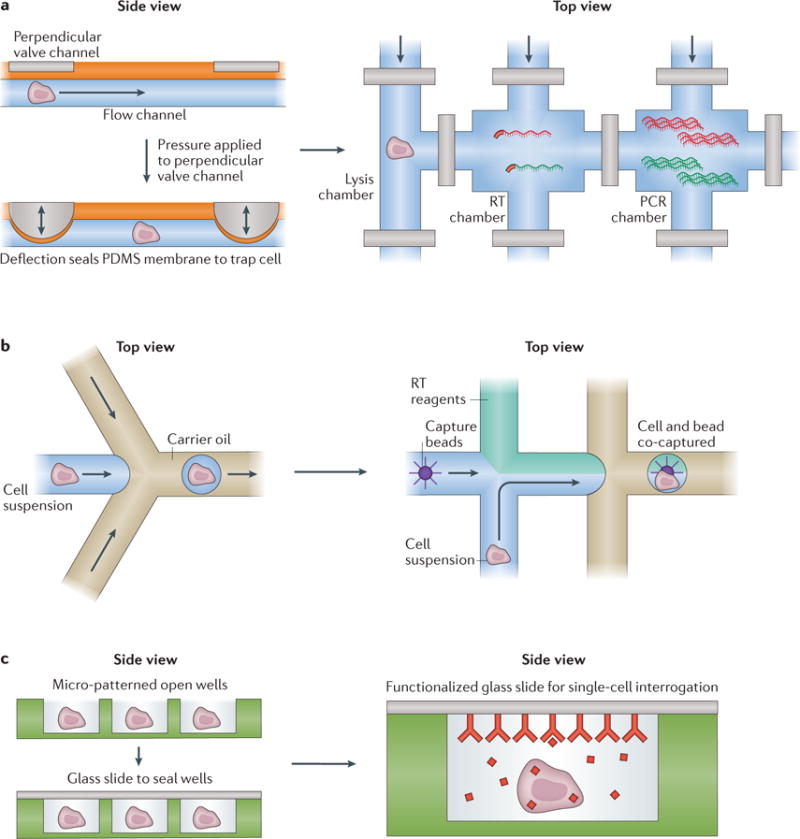
Basic microfluidic structures for processing cells (left) and example implementations (right). a | Valve-based microfluidics operate by aligning two perpendicular channels, one for cell (or cellular component) and solution flow and another for control, separated by a thin membrane. To isolate a chamber, pressure is applied to the control channel, deflecting a membrane into the flow channel to block it, and hence trap its contents. These channels can be arrayed, multiplexed and coupled to external computer-controlled pressure regulators to reliably execute complex workflows such as whole-transcriptome amplification (see, for example, REF. 3). b | In many implementations, droplet-based strategies rely on flowing single cells (or cellular components) in aqueous medium into a co-flowed oil phase. Due to pressure-driven effects, small aqueous droplets that can contain cells (or their components) are formed through encapsulation with oil, isolating each individual cell or cellular component from its peers. During or after this initial encapsulation, additional reagents, such as barcoded oligo(dT) mRNA capture beads (see, for example, REF. 31), can be incorporated or removed to enable more complex operations. c | Nanowells confine cells by gravity, and can subsequently be sealed with a membrane or glass slide to isolate single cells and their components. These cells or components can then be picked out of the wells for further processing (see, for example, REF. 5) or characterized in-well using, for example, a functionalized seal. PDMS, polydimethylsiloxane; RT, reverse transcription.
