Figure 3. Selected examples of microfluidic devices used to measure single-cell genomes and epigenomes.
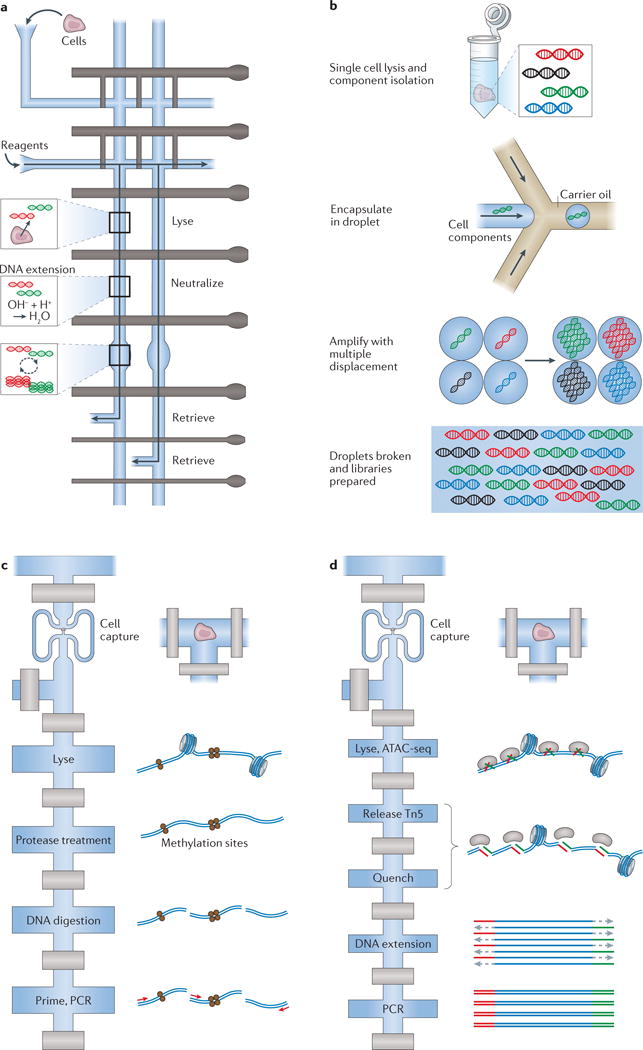
a | A diagram (top view) of the valve-based microfluidic device used by Wang et al.23 for amplification of genomic material from single cells. In the implementation, individual cells were captured and lysed, genomic DNA was then stabilized and amplified and, finally, cellular products were collected independently to ensure single-cell resolution of information. b | Emulsion multiple-displacement amplification (eMDA), as performed by Fu et al.61 (top view). Here, genomic material from single-cell lysates was encapsulated in droplets, within which MDA was performed, enabling more uniform coverage of the chromosomal profile (relative to MDA performed en masse) of the individual cell after the droplets have been broken. c | A modified diagram representing the valve-based microfluidic device implemented by Cheow et al.78 to profile the DNA methylome and transcriptome (only the methylome is shown here) of single cells (top view). d | Single-cell assay for transposase-accessible chromatin using sequencing (ATAC-seq) by Buenrostro et al.79, which uses a Tn5 transposase to identify regions of open chromatin and was performed using a commercial C1 valve-based integrated fluidic circuit from Fluidigm (top view). Part a is adapted with permission from REF. 23, Elsevier. Part c is adapted with permission from REF. 78, Macmillan Publishers. Part d is adapted with permission from REF. 79, Macmillan Publishers.
