Figure 5. Future directions and extensions for microfluidic devices.
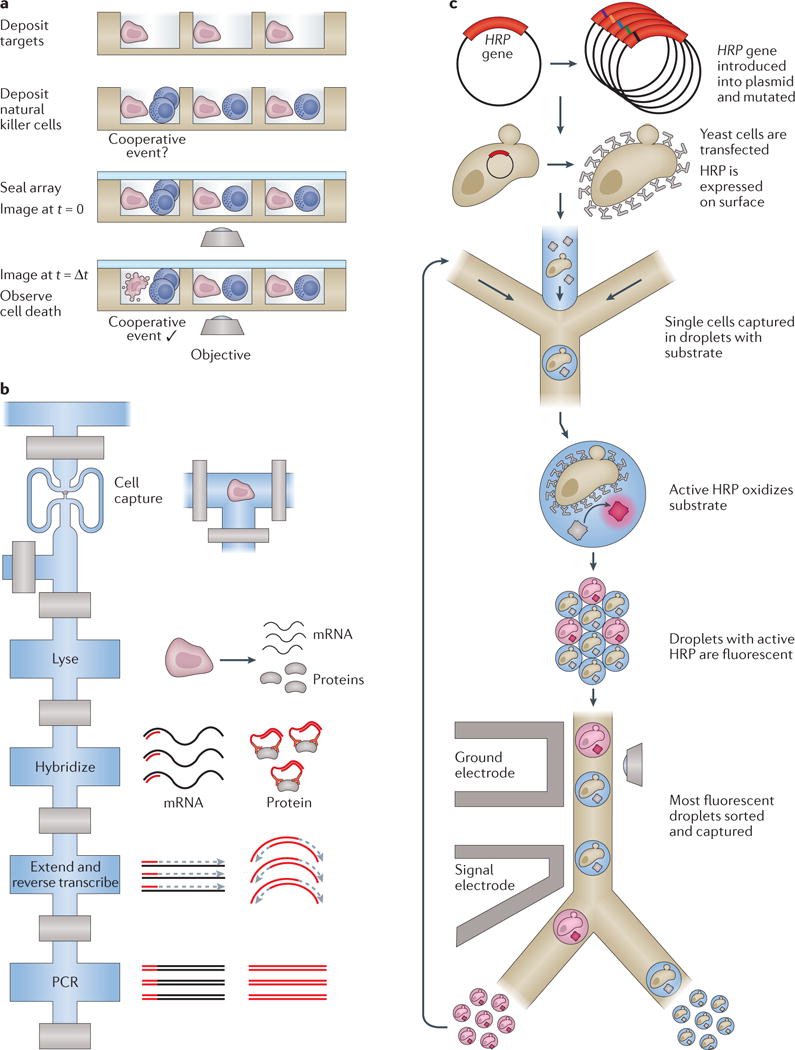
a | Multiple cells can be deposited onto well-based profiling platforms, enabling the examination of multicellular and cooperative events, such as the killing of target cells by natural killer cells by Yamanaka et al.123 (side view). b | The Fluidigm C1 valve-based integrated fluidic circuit (IFC; top view) was used to simultaneously detect several transcripts and proteins from the same single cell in a single series of reactions15. During lysis, targeted primers and complementary DNA-tagged antibodies were introduced, and reverse transcription and DNA extension reactions were carried out on both species to yield uniquely detectable, PCR-amplifiable DNAs for both cellular analytes. c | The directed evolution of enzymes in single cells can be probed with an activity assay in droplets136. Here, horseradish peroxidase (HRP) surface proteins from mutated vectors were interrogated for substrate turnover with single-cell resolution (top view). Following capture of the most active cells, the interrogation was repeated to study their evolution over time. Part a is adapted from REF. 123 with permission of The Royal Society of Chemistry. Part b is adapted with permission from REF. 15, Macmillan Publishers Limited. Part c is adapted with permission from REF. 136 (Agresti, J. J. et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl Acad. Sci. USA 107, 4004–4009; 2010).
