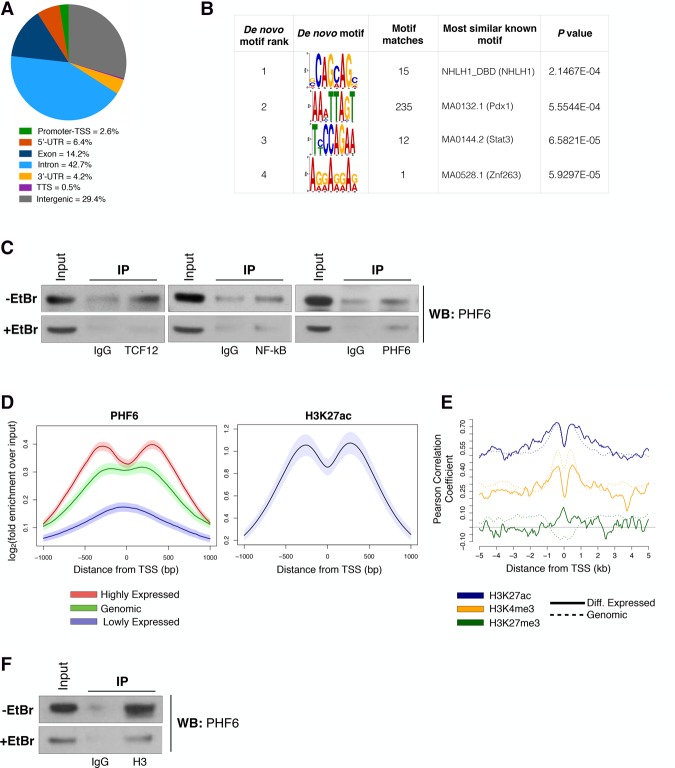
Figure 3.
PHF6 exerts transcriptional regulation by interacting with histones rather than binding sequence-specific DNA sites. (A) Pie chart showing the distribution of 77,749 PHF6-binding sites across genomic regions in B-ALL cells. (TTS) Transcription termination site; (UTR) untranslated region. (B) De novo DNA sequence motifs identified in PHF6-bound regions at promoters of differentially expressed genes with their associated P-values. Shown are sequence logos of de novo position-weight matrices found by the MEME motif discovery tool (left) or those of known transcription factors whose motifs are found to be most similar to the de novo motif discovery results by Tomtom software (right). (C) Endogenous coimmunoprecipitation (co-IP) assay of TCF12, NF-κB, and PHF6 in the absence (top) or presence (bottom) of ethidium bromide (EtBr). Input is 7% of immunoprecipitation lysate. (D, left) Metagene tracks of PHF6 ChIP-seq signal averaged over all promoter–TSS tracks grouped by relative expression levels. (Red) High; (green) genomic; (purple) low. (Right) Metagene track of H3K27ac ChIP-seq signal averaged over all promoter–TSS regions. Shaded regions around average tracks denote estimates of 95% confidence interval (CI) of the metagene average signals based on resampling. (E) Metagene tracks of PHF6 ChIP-seq signal and correlation with histone marks: H3K27ac (blue), H3K4me3 (yellow; GSE66234), H3K27me3 (green). Pearson correlation of PHF6 and histone ChIP-seq signals across 10-kb regions spanning the TSS. Differentially expressed genes (solid line) and genome-wide genes (dotted line) are shown. (F) Endogenous co-IP assay of histone H3 and PHF6 in the absence (top) or presence (bottom) of EtBr. Input is 7% of immunoprecipitation lysate.