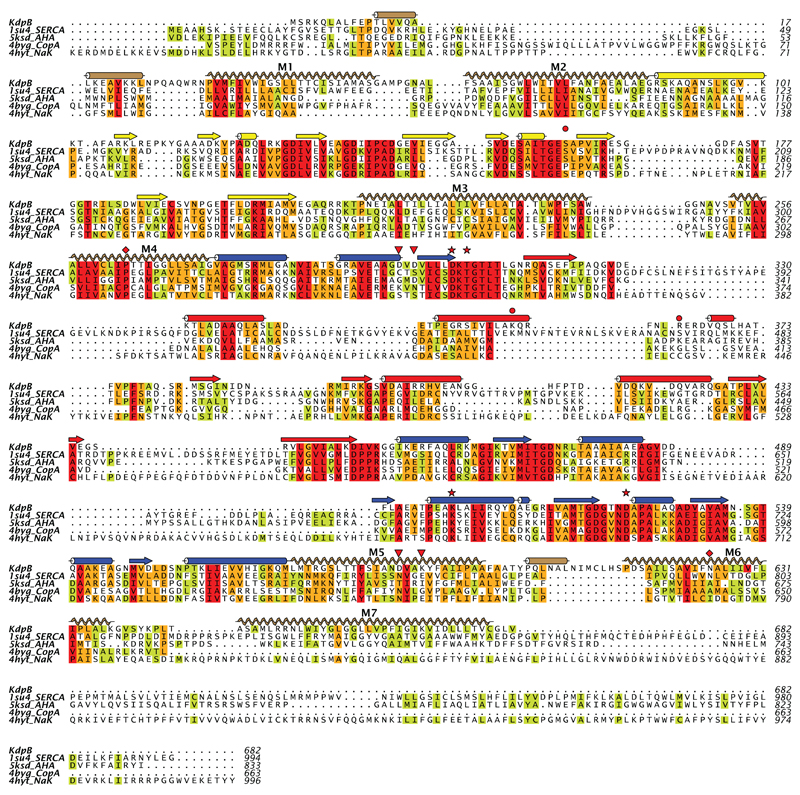
Extended Data Figure 7. Sequence alignment of KdpB with other P-type ATPases.
This structure-based alignment was done by the promals3d server using PDB depositions 1SU4 for the calcium pump (SERCA)15, 5KSD for the plasma membrane proton pump (AHA2)60, 4BYG for the copper pump (CopA)61 and 4HYT for Na,K-ATPase62. Sequence conservation is shown by the coloring scheme with red being most conserved and green least conserved. Secondary structure elements correspond to KdpB with the same coloring scheme as Fig. 1. The red stars indicate the catalytic Asp307 and other residues involved in an H-bonding network shown in Extended Data Fig. 4a (Thr309, Lys399 and Asp522). The red diamonds indicate residues at the water site along M4 and M6 shown in Extended Data Fig. 4d (Pro264 and Asn624). Red triangles indicate residues implicated in coupling with KdpA, namely Asp583 and Lys586 (Extended Data Fig. 4d) in the transmembrane domain as well as Asp300 and Asp302 that interact with cytoplasmic loops from KdpA (Fig. 3b). The red circles indicate Ser162 that is phosphorylated and Lys357/Arg363 from the N-domain that form salt bridges to the phosphate (Extended Data Fig. 5c).