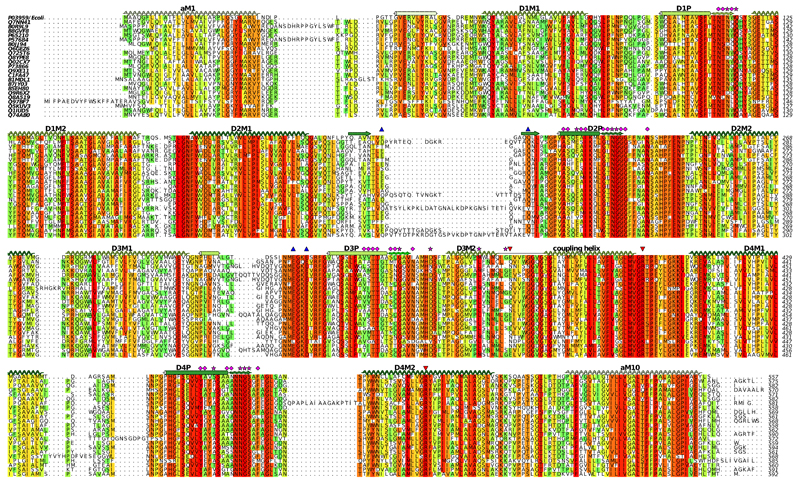
Extended Data Figure 8. Sequence alignment of KdpA from a diverse group of bacteria.
Selection of the sequences was done with the divblast server49 and alignment using clustal omega48. Secondary structure corresponds to that observed for KdpA (Fig. 1). Pink stars indicate residues reported by Buurman et al.5 and pink diamonds indicate residues reported by Dorus et al.6 to increase the apparent Kd for K+ to 0.3 mM or higher. Red triangles indicate residues implicated in coupling with KdpB, namely the Glu370 and Arg393 at the cytoplasmic end of the selectivity filter (Fig. 2c) and Arg400 that forms a salt bridge to the P-domain of KdpB (Fig. 3b). Blue triangles indicate residues forming H-bonds to the periplasmic domain of KdpC.