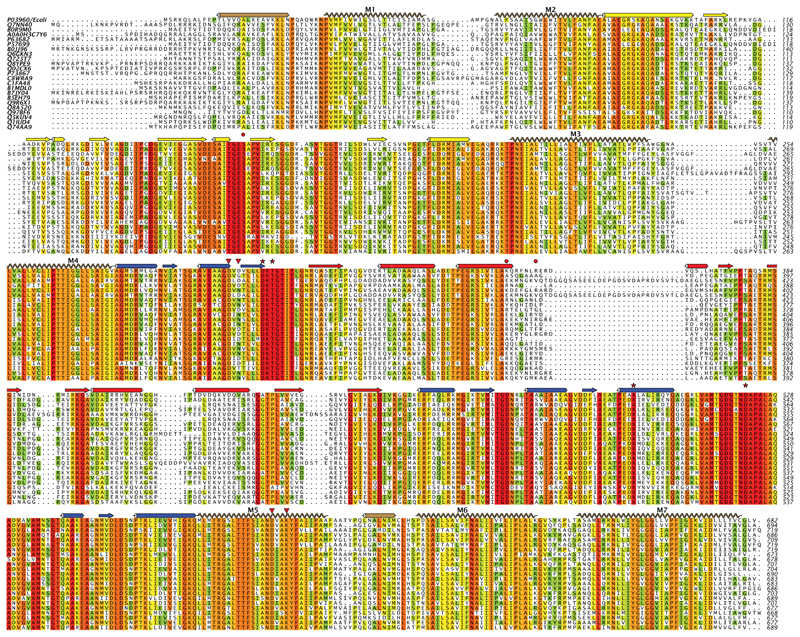
Extended Data Figure 9. Sequence alignment of KdpB from a diverse group of bacteria.
Selection of sequences was done with the divblast server49 and alignment using clustal omega48. Secondary structure corresponds to that observed for KdpB (Fig. 1). Red stars indicate residues that interact with the catalytic Asp307 in our structure. Red triangles indicate the residues that interact with cytoplasmic loops from KdpA (Asp300 and Asp302). Red circles indicate Ser162 that is phosphorylated in our structure and the residues in the N-domain that engage this phosphate in a salt bridge residues (Lys357 and Arg363).