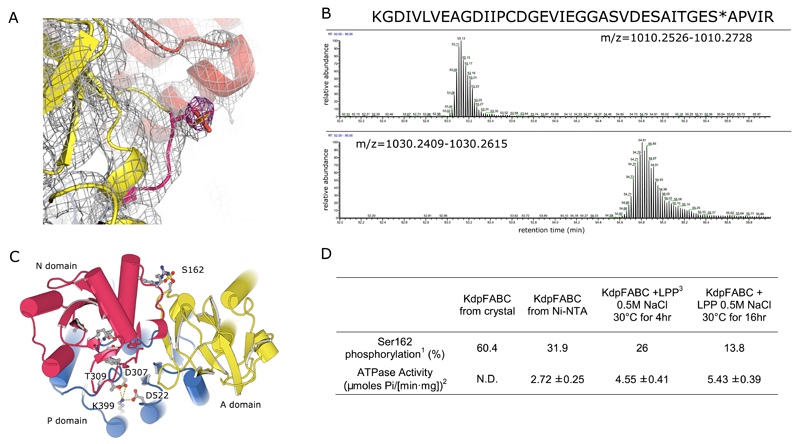
Extended Data Figure 5. Serine phosphorylation of KdpFABC.
(A) Evidence for the phosphoserine comes from extra density seen both in the 2mFo-DFc map (grey at 1.2σ) and the mFo-DFc difference density (purple at 4σ) from a refined model that lacks the phosphate group. (B) To confirm the presence of the phosphoserine, ESI-LC-MS/MS mass spectrometry was performed on KdpB isolated by SDS-PAGE from a crystal. Peptides harboring phosphorylated and unphosphorylated Ser162 were separated by FPLC and their relative abundance is represented by these elution profiles for peptides with m/z=1010.26 (unphosphorylated) or m/z=1030.25 (phosphorylated) (z=4). The sequence of these peptides, in which the cysteine was modified by iodoacetamide, is shown above, as determined by MS2. (C) The cytoplasmic domains of KdpB adopt a unique position in our structure compared to other P-type ATPases due to a salt bridge between the phosphorylated Ser162 in the A-domain (yellow) and residues Lys357 and Arg363 of the N-domain (red). This interaction likely prevents N-domain movements, and the close proximity of the P-domain to the N-domain prevents nucleotide binding, suggesting that the current structure represents an autoinhibited state. (D) The level of Ser162 phosphorylation inversely correlated with the ATPase activity of KdpFABC. The absolute level of phosphoenzyme was not determined due to a lack of a fully dephosphorylated or fully phosphorylated enzyme preparation for calibration of the mass spectrometer detection system; nevertheless, the inverse correlation between the relative levels of Ser162 phosphorylation and ATPase activity supports the inhibitory nature of this modification. Although this serine phosphorylation was observed in samples isolated directly from the bacteria, the physiological relevance is uncertain, especially given that Lys357 and Arg363 are poorly conserved amongst KdpFABC systems (Extended Data Fig. 9).
1 apparent percentage of phosphopeptide from mass spectrometry does not reflect lower detection efficiency of the phosphopeptide
2 ATPase assay was determined in the presence of 50 mM KCl
3 200 units of protein phosphatase from bacteriophage lambda (LPP) were incubated with 20 μg of KdpFABC under the conditions indicated