Abstract
The phosphatase and tensin homolog (PTEN) gene is suggested to be a dormant tumor suppressor. However, the prognostic value of the loss of PTEN expression in renal cell carcinoma (RCC) remains controversial. Therefore, we conducted a meta-analysis to evaluate the association of PTEN expression with the clinicopathological presentations and outcomes of patients with RCC through immunohistochemistry staining analysis. We systematically searched for relevant studies in PubMed, Web of Science, and Embase until March 2016. Data regarding clinical stage, pathological type, Fuhrman grade, overall survival (OS), progression-free survival (PFS), and disease-specific survival (DSS) was analyzed in the present study. In total, there were 12 studies with 2,368 patients included in this meta-analysis. The low PTEN expression in RCC was significantly associated with unfavorable DSS (HR = 1.568, 95% CI 1.015–2.242) in a random-effects model but not with OS (HR = 1.046, 95% CI 0.93–1.176) and PFS (HR = 1.244, 95% CI 0.907–1.704). Other results indicated that PTEN expression was not correlated with clinical stage, pathological type, and Fuhrman grade. This meta-analysis suggests that PTEN expression is of limited value in predicting the prognosis of patients with RCC for OS and PFS via immunohistochemistry staining analysis; and that for DSS, low PTEN expression is significantly associated with an unfavorable outcome.
Introduction
Renal cell carcinoma (RCC) is one of the most common malignancies in urological neoplasms, accounting for 2%–3% of all adult malignancies [1]. Approximately 25%–30% of metastatic lesions are detected at initial diagnosis, and metastatic RCC (mRCC) is resistant to treatment [2, 3]. Although significant advancements in the understanding of RCC and improvements in its therapeutic strategies have been achieved in the past several decades, the vast majority of patients with advanced RCC still die of their disease rather than competing causes.
The discovery and validation of biomarkers that can indicate clinical behavior and provide information about the prognosis of a given tumor are crucial to successful oncotherapy. In particular, biomarkers that can distinguish an aggressive phenotype may guide the selection of intensive treatment in clinical practice [4]. It is thought that biomarkers have the potential to inform risk stratification, estimate treatment effects, and predict prognosis.
The phosphatase and tensin homolog is a protein encoded by the PTEN gene. PTEN maps to chromosome 10q23.3 and has been thoroughly investigated as a dormant tumor suppressor [5–7]. PTEN has been demonstrated to be a component of signal transduction pathways involved in the regulation of cell growth, proliferation, and apoptosis, as well as a participant in the control of the cell cycle [8–10]. For example, PTEN was shown to antagonize the phosphoinositol-3-kinase (PI3K)/PTEN/AKT signaling pathway, which plays a crucial role in cell growth, differentiation, and survival, thereby safeguarding important cellular machineries against carcinogenesis. Thus, the loss of PTEN expression may cause the initiation of tumorigenesis [11–15].
Consequently, investigating the association between PTEN expression and the survival outcomes of patients with RCC is important and helpful in adopting personalized treatment measures. The loss of the PTEN gene, which is a master cellular regulator, is associated with tumor progression and adverse outcomes in various human cancers [16–19]. Although PTEN has been thoroughly investigated, the prognostic value of the loss of its expression in RCC remains controversial because previous reports were inconsistent [20–23]. Some studies suggested that the loss of PTEN expression demonstrated an adverse association with prognosis, but other evidence showed no correlation between the two factors [15, 23, 24]. Thus, we performed a meta-analysis to evaluate the practicability of PTEN expression as a biomarker in RCC.
Materials and methods
Search strategy selection criteria
We systematically searched for relevant studies in PubMed, Web of Science, and Embase until March 2016. “PTEN,” “phosphatase and tensin homolog,” “renal or kidney,” “neoplasm, tumor, cancer, or carcinoma,” and “prognosis, outcome, progress, mortality, and survival” were used as search terms. The start date of our searches was 2003 and the end date was 2015. The reference lists of relevant articles were examined for additional eligible studies.
The titles and abstracts of relevant studies were scrutinized to exclude irrelevant articles. The relevant articles were reviewed as full texts afterward. Studies were included in this meta-analysis if they met the following inclusion criteria: (1) investigated the clinical presentations and prognosis of patients with RCC; (2) detected PTEN protein expression intensity in immunohistochemistry staining analysis; and (3) evaluated the correlation between PTEN protein expression and clinical presentations as well as survival outcomes [overall survival (OS), disease-specific survival (DSS) and (disease-free survival (DFS)]. The exclusion criteria were: (1) papers not written in English; (2) case reports, review articles, conference abstracts, or editorial comments; and (3) determination of PTEN expression by other methods, including RT-PCR and Western blot analysis.
The selection process was performed by two reviewers independently to ensure that the studies included were eligible. When studies contained duplicated data, the study with the largest sample size was chosen. Multivariate analysis outcomes were given preference over univariate results; if no multivariate results were provided, univariate outcomes were accepted, and the survival curves in included studies were used for calculation.
Data extraction
Two investigators extracted data from included studies independently. The Newcastle–Ottawa Quality Assessment Scale (NOS) was used to assess the quality of each included study. Selection of study population, data comparability, and outcome in these cases and control groups were involved in this assessment scale. Studies with an NOS score ≥6 were defined as high-quality [25]. The following information was extracted from the included studies: the name of the first author, publication year, patient number, country, follow-up duration, antibody used, cutoff score, tumor pathological type, tumor clinical stage, Fuhrman grade, patient type (localized or metastatic RCC), DFS, DSS, OS, and PTEN expression level. In addition, we used GetData Graph Digitizer 2.26 (http://getdata-graph-digitizer.com/) to digitize and extract the survival data from a Kaplan–Meier curve given in some studies.
The cutoff value to define PTEN-low or PTEN-high expression varied across the included studies. The criteria described in the original articles were adopted.
Statistical analysis
All statistical analyses were conducted using Stata version 12.0 (StataCorp LP, TX) software. Pooled ORs (odds ratio) with 95% CI (confidence interval) were calculated to measure the correlation between PTEN expression level and clinical stage and Fuhrman grade of RCC. Pooled RRs (risk ratio) with 95% CI were adopted to evaluate the correlation between PTEN expression and survival outcomes including DFS, DSS, and OS. The Q test and I2 test were used to measure heterogeneity among studies. A fixed-effects model or a random-effects model was used depending on the heterogeneity. If the heterogeneity was significant, a random-effects model was used; otherwise, a fixed-effects model was adopted. Publication bias was evaluated by Begg’s and Egger’s tests. All P values were two-tailed, and P < 0.05 was considered statistically significant. Data concerning geographic area, staining pattern, staining cutoff, tumor pathological type, patient type (localized or metastatic), sample size, and follow-up time was used in subgroup analyses to investigate the correlation between PTEN expression level and survival outcomes. The prognostic value of PTEN expression level in terms of clinical stage, Fuhrman grade, and pathological type was measured in only two, three, and three studies, respectively; thus, pooled analyses were conducted on these factors. A sensitivity analysis was performed using the leave-one-out approach to measure the reliability of the pooled results. This systematic review and meta-analysis adhered to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guideline (Table A in S1 File).
Results
Literature search
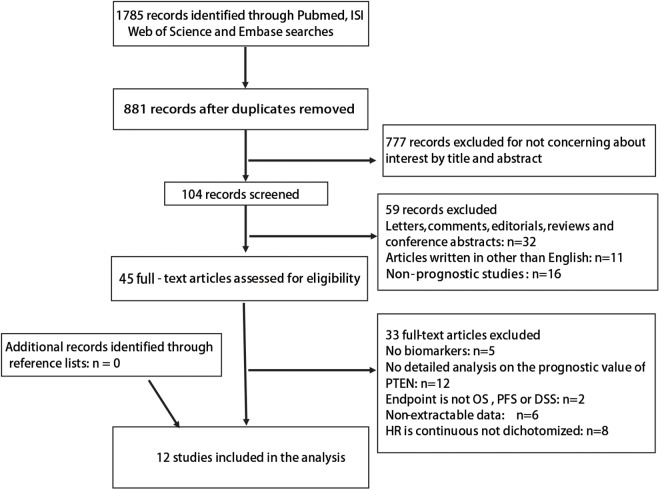
A flow chart of study selection is shown in Fig 1. A total of 1,785 records were obtained with the described search strategy. A total of 881 records were screened after exclusion of duplicates. Among these, 777 were excluded by reviewing the title and abstract. Of the remaining 104 records, 59 were excluded, including 32 letters, comments, reviews, and conference abstracts, 11 non-English articles, and 16 non-prognostic studies. The 45 remaining articles were assessed in full text. Of those, 33 articles were excluded, including: five that lacked of biomarkers; 12 missing detailed analysis on the prognostic value of PTEN; four whose endpoint was neither OS, PFS, nor DSS; six that had no extractable data; and eight in which HR was not dichotomized. A total of 12 studies with 2,368 patients were included in this meta-analysis. All the included studies investigated the association of PTEN expression with the clinicopathological presentations and outcomes of patients with RCC through immunohistochemistry staining analysis.
Fig 1. Flowchart of selection literature.
Characteristics of eligible studies
The baseline characteristics of the 12 studies are summarized in the supporting information (Table B in S1 File). Among the included studies, four were performed in America, two in Japan, two in Korea, one in China and one in Austria. The median sample size was 130 patients with RCC (range: 33–528). The median follow-up duration was 55.75 months (range: 13.8–63.5). The included patients all underwent radical or partial nephrectomy. PTEN expression was detected in immunohistochemistry staining analysis in the included studies. The survival outcome of these patients was reported in 12 articles: among them, OS was reported in five studies and PFS and DSS were presented in five articles. The clinical stage was investigated in two studies, while five studies reported the type of selected patients (localized or metastatic). Five studies investigated PTEN expression among pathological types in RCC, and three reported the Fuhrman grading. The lymph node metastasis and TNM stage were both investigated in one study. Para-carcinoma, benign renal tumors, and normal renal specimens were also evaluated by comparing three studies discussing RCC.
Effects of PTEN expression level on survival outcomes
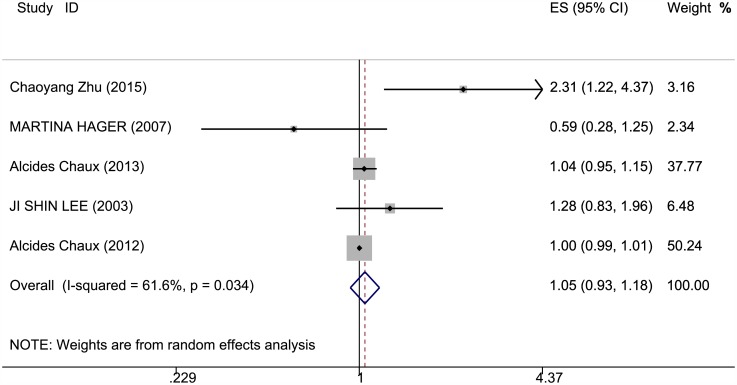
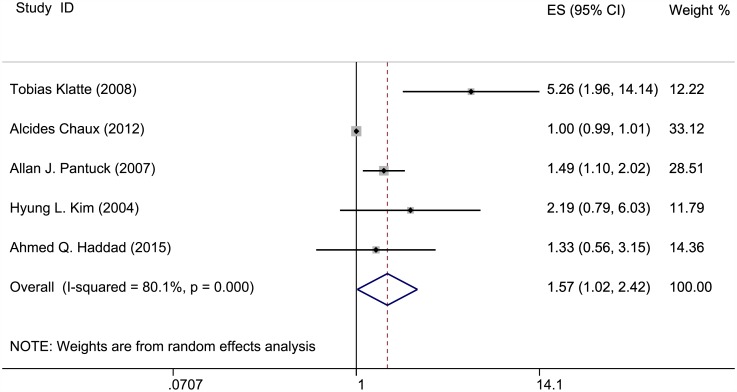
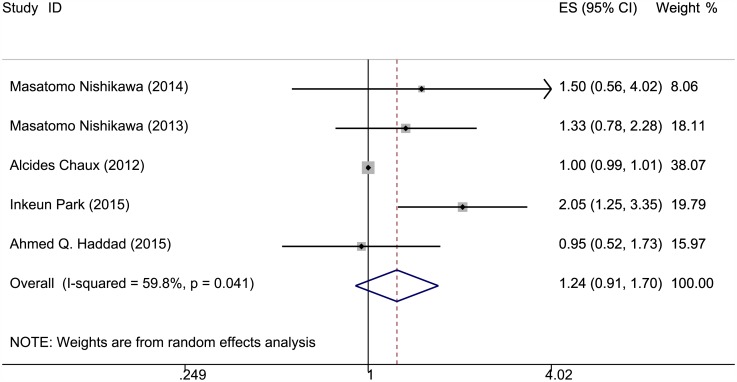
The correlation of PTEN expression and survival outcomes of patients with RCC is shown in Figs 2–4 and Table 1. OS, PFS, and DSS data were extracted from five studies. The PTEN expression level was not statistically significantly when associated with OS and PFS in the random-effects model (RR = 1.046, 95% CI = 0.93–1.176; RR = 1.244, 95% CI = 0.907–1.704); however, significant heterogeneity was observed among these studies (I2 = 61.6%, P = 0.034; I2 = 59.8%, P = 0.041). A lower PTEN expression was observed with significantly poorer DSS (RR = 1.568, 95% CI = 1.015–2.422); heterogeneity was significant in these studies as well (I2 = 80.1%, P = 0) (Table C in S1 File).
Fig 2. Meta-analysis results on PTEN expression and OS.
Forest plot of the association between PTEN expression and overall survival of patients with RCC.
Fig 4. Meta-analysis results on PTEN expression and DSS.
Forest plot of the association between PTEN expression and disease-specific survival of patients with RCC.
Table 1. Results of meta-analysis on PTEN expression.
| Overall survival (Survival vs. Death) | Progression free survival (Survival vs. Death) | Disease specific survival (Survival vs. Death) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | HR | LCI | UCI | n | HR | LCI | UCI | n | HR | LCI | UCI | |
| Overall | 5 | 1.046 | 0.93 | 1.176 | 5 | 1.244 | 0.907 | 1.704 | 5 | 1.568 | 1.015 | 2.422 |
| Geographic area | ||||||||||||
| 1. Asian | 2 | 1.637 | 0.925 | 2.897 | 3 | 1.66 | 1.181 | 2.332 | NA | NA | NA | NA |
| 2. non-Asian | 3 | 1.005 | 0.96 | 1.053 | 2 | 1 | 0.99 | 1.01 | NA | NA | NA | NA |
| Staining pattern | ||||||||||||
| 1. cytoplasm | 4 | 1.056 | 0.944 | 1.181 | NA | NA | NA | NA | NA | NA | NA | NA |
| 2.cytoplasm,nuclear | 1 | 0.59 | 0.279 | 1.247 | NA | NA | NA | NA | NA | NA | NA | NA |
| Cutoff of staining | ||||||||||||
| 1. <50% | 3 | 1.237 | 0.637 | 2.401 | 4 | 1.433 | 1.014 | 2.025 | 1 | 1 | 0.561 | 3.154 |
| 2. >=50% | 2 | 1 | 0.991 | 1.01 | 1 | 1 | 0.99 | 1.01 | 4 | 1.64 | 1 | 2.691 |
| Sample size | ||||||||||||
| 1. <100 | 4 | 1.056 | 0.944 | 1.181 | 2 | 1 | 0.99 | 1.01 | 1 | 1 | 0.99 | 1.01 |
| 2. >=100 | 1 | 0.59 | 0.279 | 1.247 | 3 | 1.41 | 0.911 | 2.181 | 4 | 1.962 | 1.15 | 3.347 |
| Follow-up | ||||||||||||
| 1. <60 | 3 | 1.314 | 0.89 | 1.94 | 2 | 1.075 | 0.644 | 1.796 | 4 | 1.962 | 1.15 | 3.347 |
| 2. >=60 | 1 | 1 | 0.99 | 1.01 | 3 | 1.331 | 0.843 | 2.102 | 1 | 1 | 0.99 | 1.01 |
| Patient type | ||||||||||||
| 1. localized | NA | NA | NA | NA | 1 | 1.33 | 0.778 | 2.274 | NA | NA | NA | NA |
| 2. metastatic | NA | NA | NA | NA | 2 | 1.926 | 1.241 | 2.989 | NA | NA | NA | NA |
| Pathological type | ||||||||||||
| 1. clear cell RCC | NA | NA | NA | NA | 1 | 0.95 | 0.521 | 1.733 | 2 | 1.639 | 0.849 | 3.166 |
| 2. other type RCC | NA | NA | NA | NA | 1 | 1 | 0.99 | 1.01 | 2 | 2.124 | 0.42 | 10.735 |
HR, hazard ratio; LCI, lower confidence interval; UCI, upper confidence interval.
Fig 3. Meta-analysis results on PTEN expression and PFS.
Forest plot of the association between PTEN expression and progression-free survival of patients with RCC.
In terms of PFS, the subgroup meta-analysis showed that the cutoff scores under 50% indicated that lower PTEN expression was associated with poorer PFS (RR = 1.43, 95% CI = 1.014–2.025, I2 = 23.3) and that lower PTEN expression was observed with poor PFS in Asian areas of the geographic area subgroup (RR = 1.66, 95% CI = 1.181–2.332, I2 = 0). Lower PTEN expression was also observed with poor PFS in patients with metastatic RCC (RR = 1.926, 95% CI = 1.241–2.989, I2 = NA). For DSS, in over 50% of the cutoff scores of the subgroup, lower PTEN expression was observed with poorer DSS (RR = 1.64, 95% CI = 1–2.691, I2 = 84.8), and a sample size of ≥100 showed a significant difference among the PTEN expression levels (RR = 1.962, 95% CI = 1.15–3.347, I2 = 52.1). The follow-up subgroup under 60 months showed poorer DSS with lower PTEN expression (RR = 1.962, 95% CI = 1.15–3.347, I2 = 52.1).
Correlations between PTEN expression and clinicopathological presentations
The associations between PTEN expression level and the clinical presentations of RCC are listed in Table 2. Respectively, three, two, and three studies were available for pooled analyses in terms of clinical stage, pathological type, and Fuhrman grade. However, no significant correlations were observed between PTEN expression levels and clinical stage, pathological type (ccRCC vs. other type), or Fuhrman grade in our meta-analysis (OR = 1.918, 95% CI = 0.502–7.336; OR = 0.781, 95% CI = 0.249–2.453; OR = 2.415, 95% CI = 0.508–9.055).
Table 2. Meta-analysis results of PTEN expression level and clinical features.
| n | OR | LCI | UCI | Heterogeneity | Publication bias | |||
|---|---|---|---|---|---|---|---|---|
| Pa | I2 (%) | Pc (Begg's test) | Pd (Egger' test) | |||||
|
Clinical stage Stage I+II vs. III+IV |
2 | 1.918 | 0.502 | 7.336 | 0.021 | 81.3 | 0.317 | NA |
|
Pathological type ccRCC vs. Other type |
3 | 0.781 | 0.249 | 2.453 | 0.017 | 75.3 | 0.602 | 0.396 |
|
Fuhrman grade I+II vs. III+IV |
3 | 2.145 | 0.508 | 9.055 | 0 | 87.6 | 0.602 | 0.084 |
OR, odds ratio; LCI, lower confidence interval; UCI, upper confidence interval; ccRCC, clear cell renal cell carcinoma.
Publication bias analysis
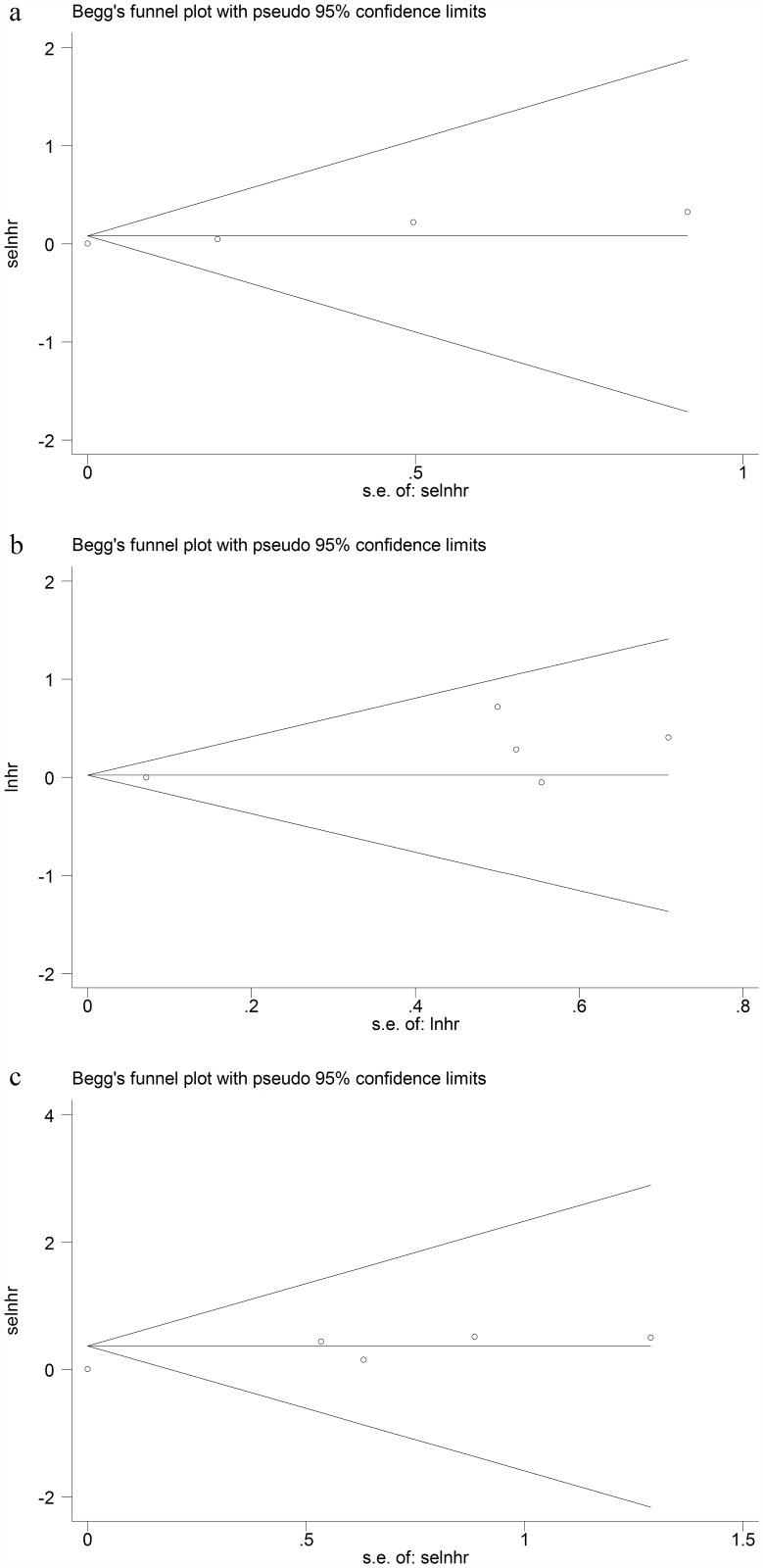
Begg’s and Egger’s tests were adopted to detect potential publication bias for OS, PFS, and DSS. No significant publication bias was observed in funnel plot asymmetry on OS, PFS and DSS. (Begg’s P value = 0.624, Egger’s P value = 0.557; Begg’s P value = 0.624, Egger’s P value = 0.158; Begg’s P value = 0.142, Egger’s P value = 0.057) (Fig 5). The results demonstrated that the included studies were almost symmetric.
Fig 5. Begg's funnel plots of publication bias for OS, PFS and DSS.
a. Begg's funnel plot for OS; b. Begg's funnel plot for PFS; c. Begg's funnel plot for DSS.
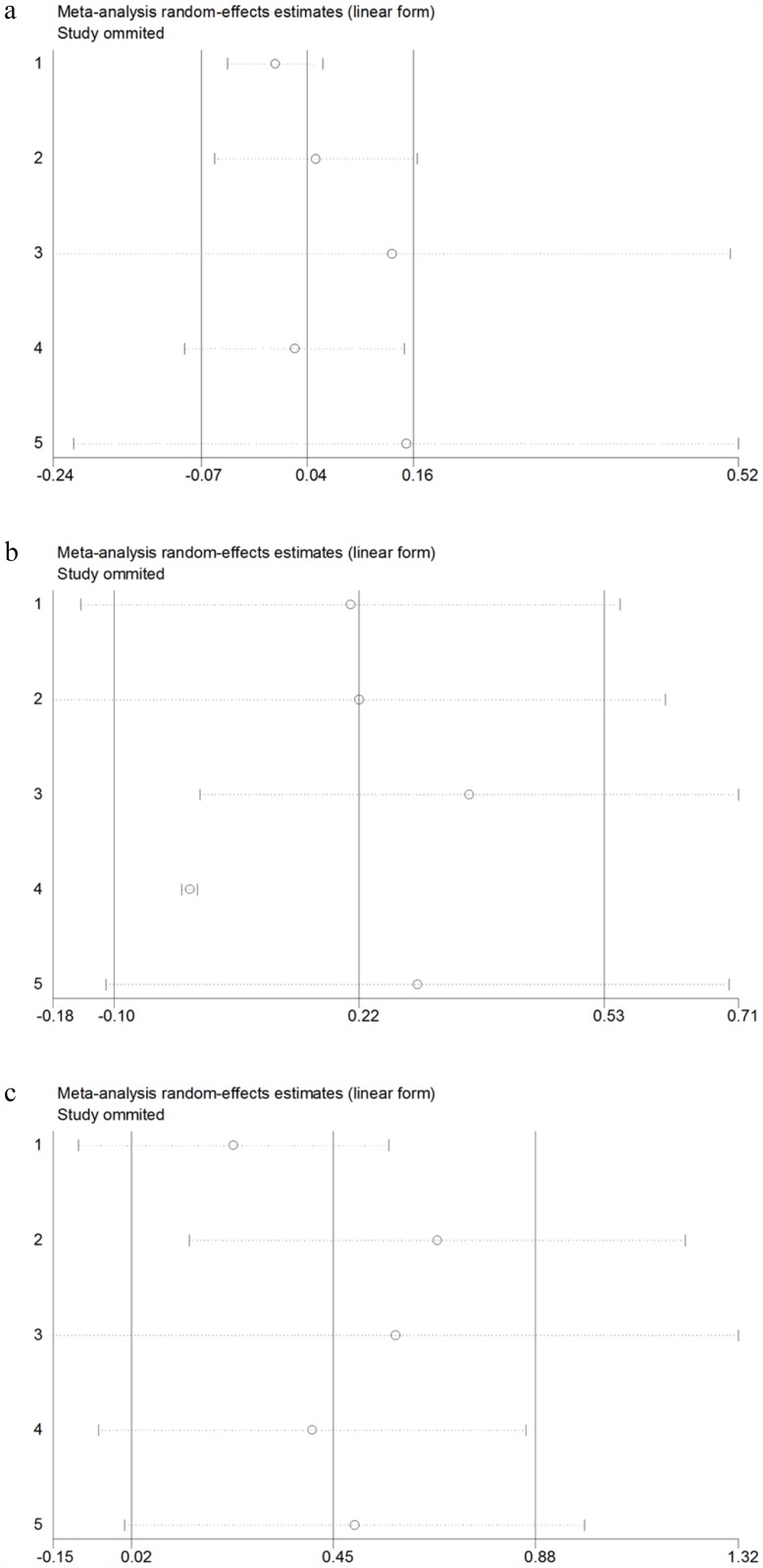
Sensitivity analysis
A sensitivity analysis was conducted using the leave-one-out approach to measure the robustness of the pooled HRs for OS, PFS, and DSS (Fig 6). The results did not differ significantly from primary results, suggesting that the pooled results were reliable.
Fig 6. Sensitivity analysis for OS, PFS and DSS.
a. Sensitivity analysis for OS; b. Sensitivity analysis for PFS; c. Sensitivity analysis for DSS.
Discussion
The present meta-analysis focused on the association between PTEN expression in immunohistochemistry staining analysis and the outcomes of patients with RCC. Overall, PTEN expression provides a significant prognostic value in DSS: patients with lower PTEN expression showed shorter DSS. No significant predictive value was detected for OS and PFS. Thus, the low PTEN expression in immunohistochemistry staining analysis presented limited value in predicting the OS and PFS. The vast majority of patients, particularly those with advanced RCC, still die of their disease rather than competing causes. Thus, PTEN expression may be used as a marker for postoperative prognosis to predict the disease specific survival. The results of subgroup analysis for OS were consistent. In terms of PFS, patients with lower PTEN expression presented significantly worse outcomes in the patients with metastatic RCC. However, for patients with localized RCC, no significant association was observed between the PTEN expression level and patient survival. No publication bias was detected. Positive PTEN was indicative of a good prognosis for receiving vascular endothelial growth factor receptor tyrosine kinase inhibitors (VEGFR TKIs) in patients with advanced RCC [26–28]. These results further demonstrated that the PTEN expression level can be a useful biomarker for treatment planning in advanced RCC.
When the cutoff of staining was below 50%, lower PTEN expression exhibited a worse outcome for PFS, whereas when the cutoff staining was over 50%, the association was absent. This result demonstrated that RCC with low PTEN expression was likely to exhibit worse prognosis. In terms of intracellular localization, PTEN expression was present predominantly in the cytoplasm of RCC cells, while only a small fraction was present in the nuclear and membrane; as such, the PTEN expression pattern of RCC is not prognostic for patient survival [24]. Brenner, et al. reported that membrane PTEN is lost in early-stage renal cell carcinogenesis and may be used as a valuable tumor marker [20]. In sample size and follow-up duration subgroup analysis, PTEN expression was correlated with the observed result of DSS. When the sample size was ≥100, the low PTEN expression presented a significant prognostic value for DSS. However, the small sample size group was not observed to exhibit worse DSS at low expression levels. The absence of significance may be attributed to the limited sample size.
Our findings indicate that PTEN expression was not correlated with clinical stage (stage I+ II vs. stage III+IV), pathological type (clear cell RCC vs. other RCC types), and Fuhrman grade (I+II vs. III+IV). This result demonstrated that the PTEN expression level is not correlated with these factors, which indicated a prognostic significance [29, 30]. Thus, the PTEN expression may not reflect the risk stratification in the clinical practice of RCC. Some studies reported that the lowest total and cytoplasmic PTEN expression levels were detected in clear-cell carcinoma and not in other subtypes [28, 31]. Some studies also showed that the PTEN expression was not associated with lymph node metastasis, tumor size, or patients’ age and gender [23, 32–34]. These results were consistent with those obtained in the current analysis.
PTEN is one of the most frequently mutated genes in human tumors, and its mutational status was identified in various cancers [35, 36]. For instance, approximately 40% of prostate cancer patients exhibited PTEN mutations, and the frequency for patients with castration-resistant prostate cancer (CRPC) was shown to be higher. PTEN loss was associated with shorter progression-free survival time [37]. In the observations of gliomas, PTEN mutations were restricted to high-grade gliomas. However, no obvious relationship was observed between the prognosis outcome and PTEN mutation status [38]. High PTEN mutation frequency was also observed in patients with breast cancer and gastric cancer [39, 40]. PTEN mutations occur in ccRCC, where they have been demonstrated to be loss-of-function mutations occurring in 1% to 5% of cases [41]. Evidence showed that the lifetime risks for RCC were projected to be approximately 30% with germline PTEN mutation [35]. Lee, et al. reported that PTEN biallelic loss was associated with an adverse ccRCC outcome, whereas monoallelic loss was not associated with poor prognosis [42]. Velickovic, et al. also reported poor prognosis in RCC patients with PTEN loss of heterozygosity [43]. de Campos, et al. reported that the deletion of PTEN in RCC was detected with a frequency of approximately 40% via fluorescent in situ hybridization and that its presence did not indicate lower survival rates [44]. Thus, PTEN mutations and loss of heterozygosity were demonstrated to play a key role in the etiology of various cancers and were associated with late-stage diseases [45]. The expression of PTEN was demonstrated to be evidently lower than that of the corresponding normal renal tissues in ccRCCs by Western blot analysis [20]. Zhu, et al. observed that PTEN expression decreased in normal renal tissues, para-carcinoma tissues, and malignant tissues [23]. These results may indicate that the loss of PTEN is involved in the entire carcinogenesis of RCC. Schneider, et al. suggested that an increased level of PTEN expression indicated a lower risk for developing metastasis, which corresponded to our results [46]. Merseburger, et al. detected no correlation between PTEN expression and patient survival [47]. Our findings showed that low PTEN expression levels corresponded to significantly shorter DSS, whereas in OS and PFS, the tendency did not reach statistical significance. Thus, PTEN as a biomarker seems to have limited usefulness in prognosis prediction.
Several limitations in our analysis should be considered. First, the retrospective studies were more susceptible to selection bias. Second, the cutoff value to define PTEN-high or PTEN-low expression varied among the original studies. Third, the number of included studies was small, which may have contributed to the negative findings in OS and PFS. Finally, the detection method of the included studies was immunohistochemistry staining analysis, which involved the use of different antibodies and processing methods that may have provided inconsistent results for prognostic markers.
Conclusion
In conclusion, the results of this meta-analysis showed that low PTEN expression was significantly associated with adverse prognosis with regard to DSS. For OS and PFS, low PTEN expression was not associated with unfavorable outcomes. Low PTEN expression presented a limited value in predicting the prognosis of patients with RCC through immunohistochemistry staining analysis. Further studies with a similar design should be conducted to confirm our results.
Supporting information
Table A in S1 File. Checklist of items to include when reporting a systematic review or meta-analysis. Table B in S1 File. Characteristics of studies included in the meta-analysis. Table C in S1 File. Heterogeneity test and publication bias analyses among studies included.
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2.Znaor A, Lortet-Tieulent J, Laversanne M, Jemal A, Bray F. International variations and trends in renal cell carcinoma incidence and mortality. European urology. 2015;67(3):519–30. doi: 10.1016/j.eururo.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 3.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. The New England journal of medicine. 2007;356(2):115–24. doi: 10.1056/NEJMoa065044 [DOI] [PubMed] [Google Scholar]
- 4.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. The New England journal of medicine. 2004;351(27):2817–26. doi: 10.1056/NEJMoa041588 [DOI] [PubMed] [Google Scholar]
- 5.Downes CP, Bennett D, McConnachie G, Leslie NR, Pass I, MacPhee C, et al. Antagonism of PI 3-kinase-dependent signalling pathways by the tumour suppressor protein, PTEN. Biochemical Society transactions. 2001;29(Pt 6):846–51. [DOI] [PubMed] [Google Scholar]
- 6.Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(8):4240–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Jiang X. PTEN: a default gate-keeping tumor suppressor with a versatile tail. Cell research. 2008;18(8):807–16. doi: 10.1038/cr.2008.83 [DOI] [PubMed] [Google Scholar]
- 8.Ortega-Molina A, Serrano M. PTEN in cancer, metabolism, and aging. Trends in endocrinology and metabolism: TEM. 2013;24(4):184–9. doi: 10.1016/j.tem.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nature reviews Molecular cell biology. 2012;13(5):283–96. doi: 10.1038/nrm3330 [DOI] [PubMed] [Google Scholar]
- 10.Yin Y, Shen WH. PTEN: a new guardian of the genome. Oncogene. 2008;27(41):5443–53. doi: 10.1038/onc.2008.241 [DOI] [PubMed] [Google Scholar]
- 11.Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer cell. 2004;6(2):117–27. doi: 10.1016/j.ccr.2004.06.022 [DOI] [PubMed] [Google Scholar]
- 12.Weng LP, Smith WM, Dahia PL, Ziebold U, Gil E, Lees JA, et al. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer research. 1999;59(22):5808–14. [PubMed] [Google Scholar]
- 13.Vazquez F, Sellers WR. The PTEN tumor suppressor protein: an antagonist of phosphoinositide 3-kinase signaling. Biochimica et biophysica acta. 2000;1470(1):M21–35. [DOI] [PubMed] [Google Scholar]
- 14.Sun L, Liu J, Yuan Q, Xing C, Yuan Y. Association between PTEN Gene IVS4 polymorphism and risk of cancer: a meta-analysis. PloS one. 2014;9(6):e98851 doi: 10.1371/journal.pone.0098851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HL, Seligson D, Liu X, Janzen N, Bui MH, Yu H, et al. Using protein expressions to predict survival in clear cell renal carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10(16):5464–71. [DOI] [PubMed] [Google Scholar]
- 16.Tachibana M, Shibakita M, Ohno S, Kinugasa S, Yoshimura H, Ueda S, et al. Expression and prognostic significance of PTEN product protein in patients with esophageal squamous cell carcinoma. Cancer. 2002;94(7):1955–60. [DOI] [PubMed] [Google Scholar]
- 17.Bose S, Crane A, Hibshoosh H, Mansukhani M, Sandweis L, Parsons R. Reduced expression of PTEN correlates with breast cancer progression. Human pathology. 2002;33(4):405–9. [DOI] [PubMed] [Google Scholar]
- 18.Lee JI, Soria JC, Hassan KA, El-Naggar AK, Tang X, Liu DD, et al. Loss of PTEN expression as a prognostic marker for tongue cancer. Archives of otolaryngology—head & neck surgery. 2001;127(12):1441–5. [DOI] [PubMed] [Google Scholar]
- 19.Salvesen HB, Stefansson I, Kalvenes MB, Das S, Akslen LA. Loss of PTEN expression is associated with metastatic disease in patients with endometrial carcinoma. Cancer. 2002;94(8):2185–91. doi: 10.1002/cncr.10434 [DOI] [PubMed] [Google Scholar]
- 20.Brenner W, Farber G, Herget T, Lehr HA, Hengstler JG, Thuroff JW. Loss of tumor suppressor protein PTEN during renal carcinogenesis. International journal of cancer. 2002;99(1):53–7. [DOI] [PubMed] [Google Scholar]
- 21.Nishikawa M, Miyake H, Harada K, Fujisawa M. Expression of molecular markers associated with the mammalian target of rapamycin pathway in nonmetastatic renal cell carcinoma: Effect on prognostic outcomes following radical nephrectomy. Urologic oncology. 2014;32(1):49.e15–21. [DOI] [PubMed] [Google Scholar]
- 22.Kim HL, Seligson D, Liu X, Janzen N, Bui MH, Yu H, et al. Using tumor markers to predict the survival of patients with metastatic renal cell carcinoma. The Journal of urology. 2005;173(5):1496–501. doi: 10.1097/01.ju.0000154351.37249.f0 [DOI] [PubMed] [Google Scholar]
- 23.Zhu C, Wei J, Tian X, Li Y, Li X. Prognostic role of PPAR-gamma and PTEN in the renal cell carcinoma. International journal of clinical and experimental pathology. 2015;8(10):12668–77. [PMC free article] [PubMed] [Google Scholar]
- 24.Hager M, Haufe H, Kemmerling R, Mikuz G, Kolbitsch C, Moser PL. PTEN expression in renal cell carcinoma and oncocytoma and prognosis. Pathology. 2007;39(5):482–5. doi: 10.1080/00313020701570012 [DOI] [PubMed] [Google Scholar]
- 25.Wells GA SB OCD, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in metaanalyses. http://wwwohrica/programs/clinical_epidemiology/oxfordasp. 2008.
- 26.Park I, Cho YM, Lee JL, Ahn JH, Lee DH. Prognostic tissue biomarker exploration for patients with metastatic renal cell carcinoma receiving vascular endothelial growth factor receptor tyrosine kinase inhibitors. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(4):4919–27. [DOI] [PubMed] [Google Scholar]
- 27.Muriel Lopez C, Esteban E, Berros JP, Pardo P, Astudillo A, Izquierdo M, et al. Prognostic factors in patients with advanced renal cell carcinoma. Clinical genitourinary cancer. 2012;10(4):262–70. doi: 10.1016/j.clgc.2012.06.005 [DOI] [PubMed] [Google Scholar]
- 28.Pantuck AJ, Seligson DB, Klatte T, Yu H, Leppert JT, Moore L, et al. Prognostic relevance of the mTOR pathway in renal cell carcinoma: implications for molecular patient selection for targeted therapy. Cancer. 2007;109(11):2257–67. doi: 10.1002/cncr.22677 [DOI] [PubMed] [Google Scholar]
- 29.Delahunt B, Cheville JC, Martignoni G, Humphrey PA, Magi-Galluzzi C, McKenney J, et al. The International Society of Urological Pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. The American journal of surgical pathology. 2013;37(10):1490–504. [DOI] [PubMed] [Google Scholar]
- 30.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. The American journal of surgical pathology. 1982;6(7):655–63. [DOI] [PubMed] [Google Scholar]
- 31.Hara S, Oya M, Mizuno R, Horiguchi A, Marumo K, Murai M. Akt activation in renal cell carcinoma: contribution of a decreased PTEN expression and the induction of apoptosis by an Akt inhibitor. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2005;16(6):928–33. [DOI] [PubMed] [Google Scholar]
- 32.Shin Lee J, Seok Kim H, Bok Kim Y, Cheol Lee M, Soo Park C. Expression of PTEN in renal cell carcinoma and its relation to tumor behavior and growth. Journal of surgical oncology. 2003;84(3):166–72. doi: 10.1002/jso.10302 [DOI] [PubMed] [Google Scholar]
- 33.Chaux A, Schultz L, Albadine R, Hicks J, Kim JJ, Allaf ME, et al. Immunoexpression status and prognostic value of mammalian target of rapamycin and hypoxia-induced pathway members in papillary cell renal cell carcinomas. Human pathology. 2012;43(12):2129–37. doi: 10.1016/j.humpath.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaux A, Albadine R, Schultz L, Hicks J, Carducci MA, Argani P, et al. Dysregulation of the mammalian target of rapamycin pathway in chromophobe renal cell carcinomas. Human pathology. 2013;44(10):2323–30. doi: 10.1016/j.humpath.2013.05.014 [DOI] [PubMed] [Google Scholar]
- 35.Tan MH, Mester JL, Ngeow J, Rybicki LA, Orloff MS, Eng C. Lifetime cancer risks in individuals with germline PTEN mutations. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18(2):400–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simpson L, Parsons R. PTEN: life as a tumor suppressor. Experimental cell research. 2001;264(1):29–41. doi: 10.1006/excr.2000.5130 [DOI] [PubMed] [Google Scholar]
- 37.Dong JT. Prevalent mutations in prostate cancer. Journal of cellular biochemistry. 2006;97(3):433–47. doi: 10.1002/jcb.20696 [DOI] [PubMed] [Google Scholar]
- 38.Rasheed BK, Stenzel TT, McLendon RE, Parsons R, Friedman AH, Friedman HS, et al. PTEN gene mutations are seen in high-grade but not in low-grade gliomas. Cancer research. 1997;57(19):4187–90. [PubMed] [Google Scholar]
- 39.Li YL, Tian Z, Wu DY, Fu BY, Xin Y. Loss of heterozygosity on 10q23.3 and mutation of tumor suppressor gene PTEN in gastric cancer and precancerous lesions. World journal of gastroenterology. 2005;11(2):285–8. doi: 10.3748/wjg.v11.i2.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kechagioglou P, Papi RM, Provatopoulou X, Kalogera E, Papadimitriou E, Grigoropoulos P, et al. Tumor suppressor PTEN in breast cancer: heterozygosity, mutations and protein expression. Anticancer research. 2014;34(3):1387–400. [PubMed] [Google Scholar]
- 41.Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(7456):43–9. doi: 10.1038/nature12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HJ, Lee HY, Lee JH, Lee H, Kang G, Song JS, et al. Prognostic significance of biallelic loss of PTEN in clear cell renal cell carcinoma. The Journal of urology. 2014;192(3):940–6. doi: 10.1016/j.juro.2014.03.097 [DOI] [PubMed] [Google Scholar]
- 43.Velickovic M, Delahunt B, McIver B, Grebe SK. Intragenic PTEN/MMAC1 loss of heterozygosity in conventional (clear-cell) renal cell carcinoma is associated with poor patient prognosis. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2002;15(5):479–85. [DOI] [PubMed] [Google Scholar]
- 44.de Campos EC, da Fonseca FP, Zequ Sde C, Guimaraes GC, Soares FA, Lopes A. Analysis of PTEN gene by fluorescent in situ hybridization in renal cell carcinoma. Revista do Colegio Brasileiro de Cirurgioes. 2013;40(6):471–5. [DOI] [PubMed] [Google Scholar]
- 45.Minami A, Nakanishi A, Ogura Y, Kitagishi Y, Matsuda S. Connection between Tumor Suppressor BRCA1 and PTEN in Damaged DNA Repair. Frontiers in oncology. 2014;4:318 doi: 10.3389/fonc.2014.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider E, Keppler R, Prawitt D, Steinwender C, Roos FC, Thuroff JW, et al. Migration of renal tumor cells depends on dephosphorylation of Shc by PTEN. International journal of oncology. 2011;38(3):823–31. doi: 10.3892/ijo.2010.893 [DOI] [PubMed] [Google Scholar]
- 47.Merseburger AS, Hennenlotter J, Kuehs U, Simon P, Kruck S, Koch E, et al. Activation of PI3K is associated with reduced survival in renal cell carcinoma. Urologia internationalis. 2008;80(4):372–7. doi: 10.1159/000132694 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A in S1 File. Checklist of items to include when reporting a systematic review or meta-analysis. Table B in S1 File. Characteristics of studies included in the meta-analysis. Table C in S1 File. Heterogeneity test and publication bias analyses among studies included.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.