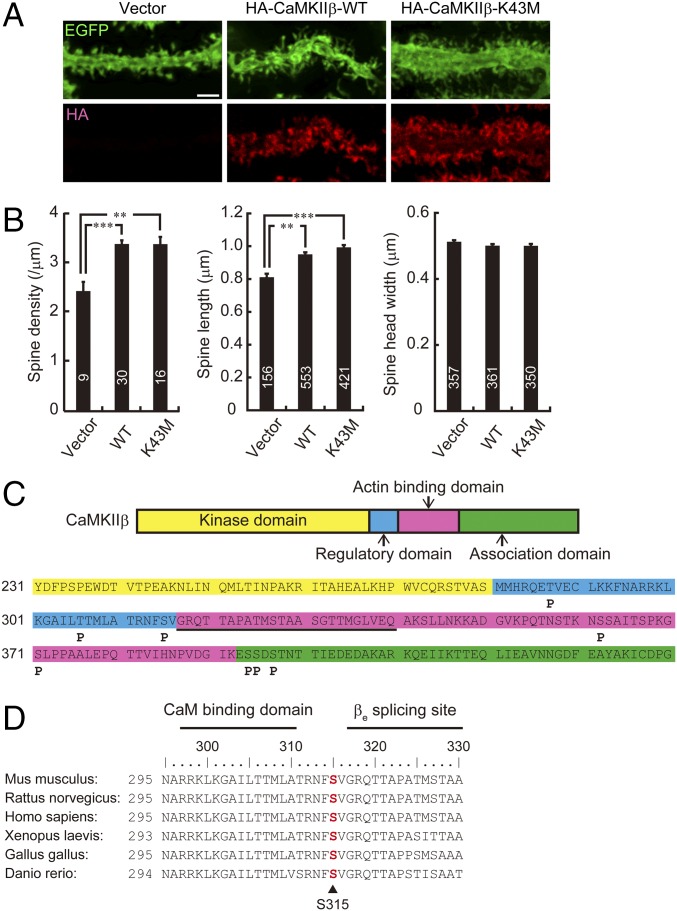
Fig. 1.
CaMKIIβ controls spine morphology and is phosphorylated at S315 in Purkinje cells. (A) Effects of overexpression of WT or K43M mutant form of HA-tagged CaMKIIβ on Purkinje cell dendritic spine morphology in cerebellar slice cultures. Cerebellar slices were subjected to biolistic cotransfection with HA-CaMKIIβ and EGFP, cultured for 2 d, and immunostained with anti-EGFP and HA antibodies. Representative images of distal dendrites of Purkinje cells are shown. (Scale bar, 2 μm.) (B) Quantitative analysis of spine density along distal dendrites (Left), spine length (Center), and spine head width (Right) of EGFP+ Purkinje cells in A. **P < 0.01, ***P < 0.0001, one-way ANOVA with Bonferroni’s test for multiple comparisons. (C) Summary of in vivo phosphorylation sites of CaMKIIβ in Purkinje cells. Phosphorylation sites are depicted as “P.” The critical F-actin binding region (317–340 aa) is underlined. Detailed information is provided in Fig. S1. (D) Conservation of S315 (in red) in CaMKIIβ among species. Calmodulin (CaM) binding domain and βe splicing site of CaMKIIβ are also shown. Each amino acid sequence is from accession numbers NP_031621.3 (Mus musculus), NP_068507.2 (Rattus norvegicus), NP_742075.1 (Homo sapiens), NP_001084063.1 (Xenopus laevis), NP_989625.1 (Gallus gallus), or XP_009303979.1 (Danio rerio). The numbers of neurons or spines are indicated in each graph.