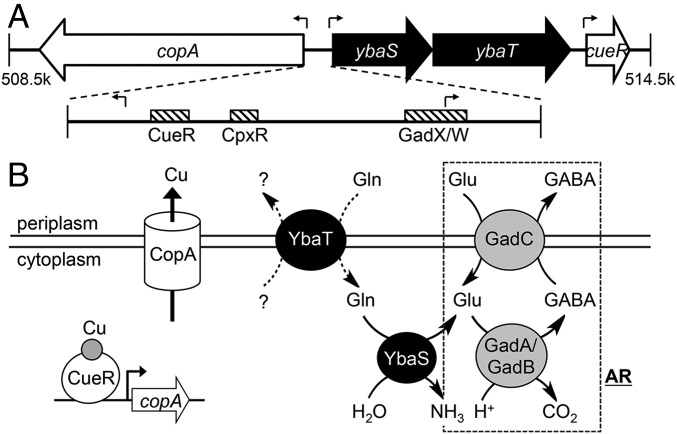
Fig. 1.
Clustering of Cu tolerance and acid tolerance genes in E. coli. (A) Genomic context of copA and cueR in E. coli. Figure shows the approximate locations of genes on the reference genome (RefSeq NC_000913.3). Block arrows represent directions of ORFs. Transcription start sites are indicated by bent line arrows. Striped boxes represent binding sites for transcription factors (CueR, accttccagcaaggggaaggt; CpxR, gtaaaagtccgtaaa; and GadX/W, taaatcaggatgcctgaaaatcggcaccggggtg). (B) Biochemical function of CopA, CueR, YbaS, and YbaT. CueR is a Cu sensor, whereas CopA is a Cu efflux pump. Both constitute the central mechanism for Cu tolerance in E. coli. YbaS is a glutaminase, whereas YbaT is a putative Gln-importing permease. The dashed box shows components of the Glu-dependent acid resistance system (AR), namely the glutamate decarboxylases GadA and GadB, as well as the Glu/GABA-antiporter GadC. Both YbaS and YbaT are thought to support the function of this AR.