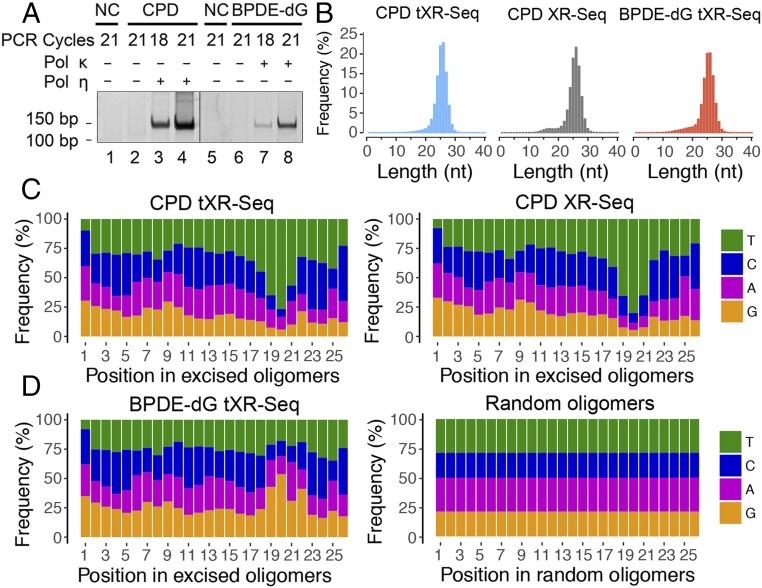
Fig. 2.
Analyses of length distribution and single-nucleotide frequencies for the excised oligomers obtained from tXR-seq and conventional XR-seq. (A) dsDNA library for tXR-seq analyzed by 10% native polyacrylamide gel electrophoresis. Primer extension products (1%) were amplified by PCR using the indicated cycles. NC, nontemplate control. (B) Length distribution of excised oligomers from CPD and BPDE-dG tXR-seq (∼20 million each) and CPD XR-seq 6 (∼10 million) after removal of adaptors and duplicate reads. (C) Single-nucleotide frequencies for 26-mers obtained by CPD tXR-seq (∼13 million) and conventional CPD XR-seq 6 (∼10 million). (D) Single-nucleotide frequencies for the 26-mers from BPDE-dG tXR-seq (∼12 million) along with single-nucleotide distribution of randomly selected 26-mers from the human genome (10 million).