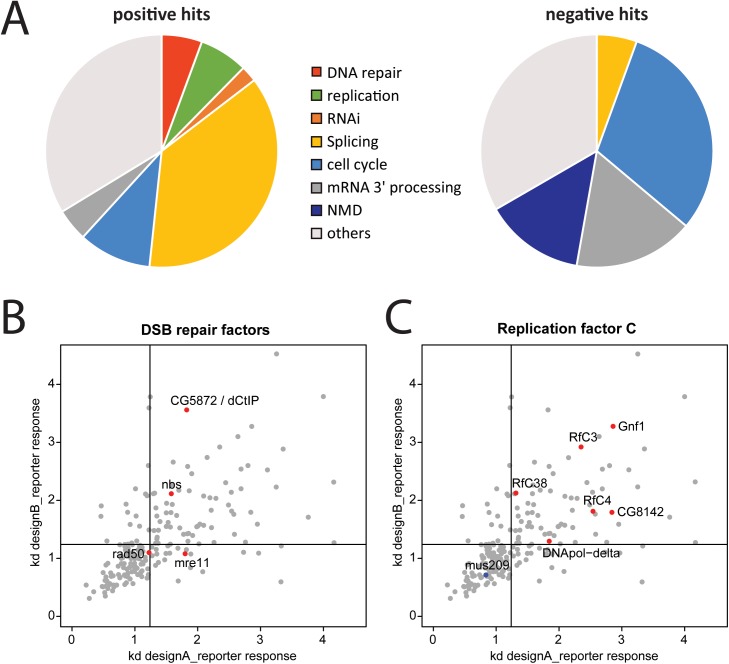
Fig 1. Analysis of the screening data for siRNA induction at a DNA double-strand break.
(A) Pie-charts of selected gene ontology terms among the 89 positive and 36 negative candidates obtained after screening and validation; the genes were analyzed with GOrilla [55]. (B) Specific labeling of the MRE11-Rad50-Nbs1 (MRN) complex in the context of the validation dataset. The putative Drosophila CtIP homolog, CG5872, was also recovered and confirmed in our screen. (C) Specific labeling of the replication factor C (RfC) subunits and DNA polymerase δ in the context of the validation dataset. Note that the Drosophila PCNA homolog, mus209, could not be confirmed as a negative hit in our validation experiments. For (A) and (C), the threshold lines were set at the mean plus 1x standard deviation of our negative controls (knock-down of GFP/DsRed).