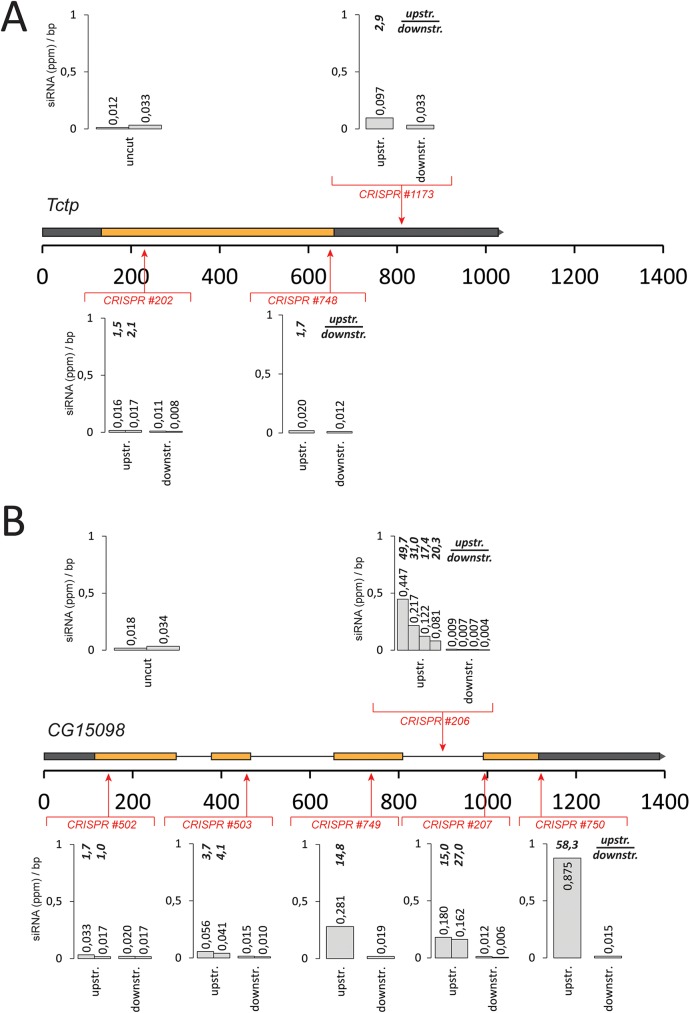
Fig 3. Correlation between gene structure and efficiency of DSB-induced siRNA formation.
We calculated the normalized read frequency (reads per million) and determined the per base average for the region between the transcription start site up to the cut (“upstream”, excluding the region where reads derived from the sgRNA itself map) and for the region between the cut until the annotated transcript 3’-end (“downstream”). Detailed traces of siRNAs mapping to the respective loci are provided in S5 Fig (TCTP) and 6 (CG15098). (A) cas9-CRISPR cuts in the intronless tctp gene result in barely any production of siRNAs. (B) Cuts in the intron-containing gene CG15098 can efficiently produce DNA-damage dependent siRNAs, provided that the cut is sufficiently downstream of an intron.