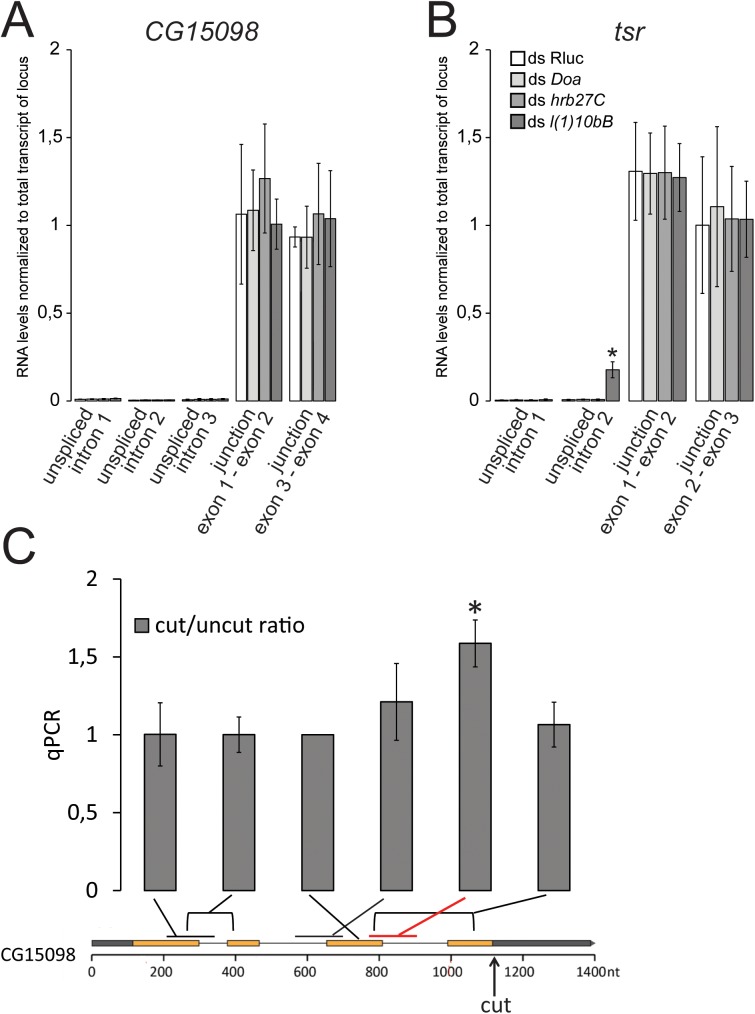
Fig 4. Knock-down of the screened splicing factors does not lead to a major decrease in overall splicing efficiency.
(A) Following knock-down of a selected set of identified splicing factors, the levels of unspliced pre-mRNA were determined with amplicons that contained the intron-exon junction, the spliced messages were specifically amplified with amplicons where one primer spanned the exon-exon junction. An amplicon confined within an exon was used to normalize to the total amount of transcript produced at the locus. The values shown are the mean ± SD of three biological replicates.No increase of pre-mRNA levels was detected at any of the three introns in our CG15098 model locus and the levels of spliced transcript were comparable with the total transcript levels. (B) For the second intron of the tsr gene, we detected an increase of unspliced pre-mRNA after knock-down of l(1)10Bb. Nonetheless, the levels of spliced message were comparable to the total transcript levels, indicating that even in this case the majority of transcripts are still spliced. * p = 0.023, Student’s t-test (two-sided, n = 3 biological replicates) (C) Induction of a downstream DNA break slows CG15098 transcript maturation. We induced cleavage at the CG15098 lcous or in the TCTP gene as a control, isolated total RNA and performed qPCR analysis with random primed cDNA. The nascent RNA levels are much lower than those of mature CG15098 (see part A), but calculating the ratio of cut vs. uncut samples displays only the DNA-break induced changes. We detected a ~1.6-fold increase of nascent RNA directly upstream of the break (p<0.01, student’s t-test, 3 biological replicates) and there may be a trend towards slightly increased levels of nascent RNA further upstream at the second intron. We did not observe any changes in the level of mature CG15098 mRNA, which was to be expected given its far greater abundance and presumably slower turnover.