Figure 2.
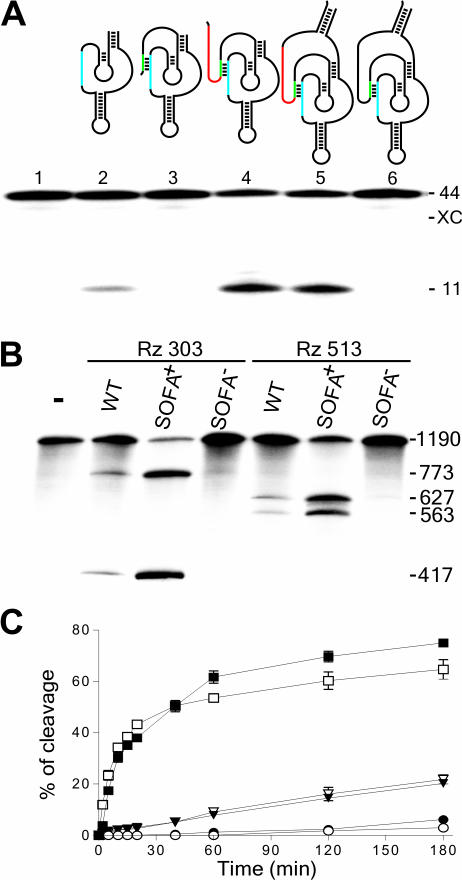
Rational design and proof-of-concept of the SOFA-ribozyme. (A) Schematic representation and typical autoradiogram of a denaturing 6% PAGE gel used to analyze the cleavage reactions of various versions of ribozyme. The P1 stem of the ribozyme (Rz), the blocker and the biosensor (BS) are in blue, green and red, respectively. The length of the bands, in nucleotides, is shown adjacent to the gel. The control (−) was performed in the absence of ribozyme (lane1), while lane 2 is in the presence of the original δRz. Lanes 3–5 are ribozymes progressively extended by the addition of the blocker (lane 3), the biosensor (lane 4) and the stabilizer (lane 5). Lane 6 is a SOFA−-ribozyme that consists of a ribozyme harboring an inappropriate biosensor sequence. (B) Typical autoradiogram of a denaturing 6% PAGE gel used to analyzing the cleavage reaction of the HBV-derived target by the original- (WT), SOFA-δRz-303 and -513 ribozymes. The incubation time was 3 h at 37°C. The length of the bands in nucleotides is shown adjacent to the gel. The control (−) was performed in the absence of ribozyme, while SOFA+ and SOFA− indicates SOFA harboring either the appropriate or inappropriate biosensor sequences, respectively. (C) Graphical representations of time courses for the cleavage reactions of SOFA+ (squares), SOFA− (circles) and the original (inversed triangles) ribozyme versions of δRz-303 (filled symbols) and δRz-513 (open symbols). Incubations were performed at 37°C and aliquots were recovered at 0, 2, 5, 10, 15, 20, 40, 60, 120 and 180 min.