Fig. 11.
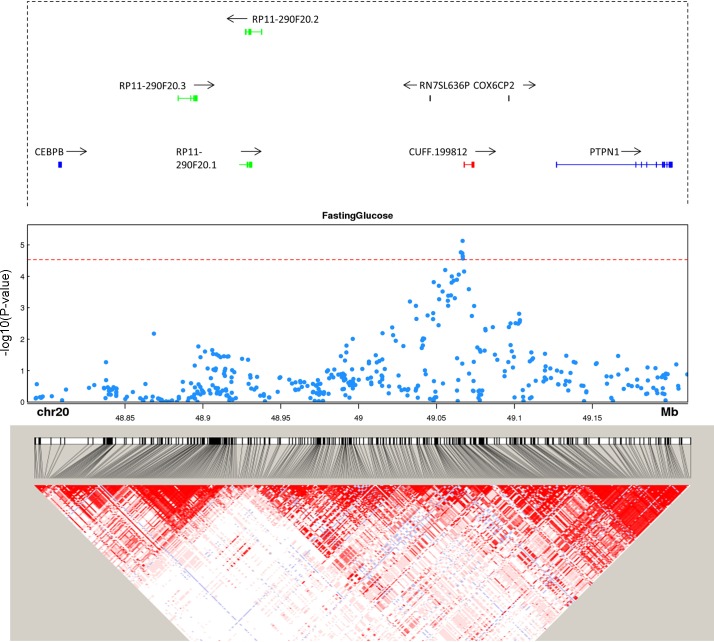
Trait-associated single nucleotide polymorphisms (SNPs) overlap unannotated lincRNA CUFF.199812 at genome-wide association study (GWAS) loci. Gene annotation, trait-associated SNPs, and linkage disequilibrium (LD) structure are shown for CUFF.199812. Genes from GENCODE v19 are plotted in each locus. Protein coding genes are shown in blue, unannotated lincRNAs in red, other lincRNAs in green. All other genes, e.g., COX6CP2, are shown in black. P value cut-off is corrected for the number of SNPs mapped to lincRNAs. LD heat map color scheme is the default D′/LOD scheme in Haploview.