Abstract
Nonalcoholic fatty liver disease (NAFLD) is the most common liver disease worldwide, yet the pathogenesis of NAFLD is only partially understood. Here, we investigated the role of the gut bacteria in NAFLD by stimulating the gut bacteria via feeding mice the fermentable dietary fiber, guar gum (GG), and suppressing the gut bacteria via chronic oral administration of antibiotics. GG feeding profoundly altered the gut microbiota composition, in parallel with reduced diet-induced obesity and improved glucose tolerance. Strikingly, despite reducing adipose tissue mass and inflammation, GG enhanced hepatic inflammation and fibrosis, concurrent with markedly elevated plasma and hepatic bile acid levels. Consistent with a role of elevated bile acids in the liver phenotype, treatment of mice with taurocholic acid stimulated hepatic inflammation and fibrosis. In contrast to GG, chronic oral administration of antibiotics effectively suppressed the gut bacteria, decreased portal secondary bile acid levels, and attenuated hepatic inflammation and fibrosis. Neither GG nor antibiotics influenced plasma lipopolysaccharide levels. In conclusion, our data indicate a causal link between changes in gut microbiota and hepatic inflammation and fibrosis in a mouse model of NAFLD, possibly via alterations in bile acids.
Keywords: antibiotics, hepatic fibrosis, hepatic inflammation, obesity, inflammation, intestine
The worldwide epidemic of obesity is the primary driver for the global increase in the prevalence of type 2 diabetes and nonalcoholic fatty liver disease (NAFLD) (1). While caloric overconsumption and the associated positive energy balance are the sine qua non of obesity development, emerging evidence implicates gut microbes in the promotion of obesity and related metabolic disturbances (2–5).
The human intestine contains a variety of microbiota, mainly consisting of bacteria and complemented by other microorganisms, such as fungi, protozoa, and viruses. The gut microbiota form a mutualistic relationship with the host and have an important role in host health. Besides protecting the host against invading pathogenic microorganisms, intestinal bacteria facilitate the digestion of otherwise indigestible carbohydrates, produce essential vitamins, stimulate the development of the immune system, and maintain tissue homeostasis (6, 7). In recent years, the gut microbiota have increasingly been connected with a number of diseases, including irritable bowel syndrome, Crohn’s disease, obesity, type 2 diabetes, atherosclerosis, and NAFLD (5, 8–10).
NAFLD describes a spectrum of related liver diseases ranging from simple steatosis to nonalcoholic steatohepatitis (NASH), liver fibrosis, and cirrhosis (11). Although the exact pathogenesis of NAFLD is unknown, multiple factors likely contribute to the progression of NAFLD, including genetic predisposition, lipid overload, and inflammatory insults. Indeed, it is believed that inflammatory mediators released locally or derived from tissues, such as the intestine and adipose tissue, play an important role in the progression of steatosis to NASH (12, 13).
Recent studies have raised the possibility that NAFLD may be related to disturbances in the gut microbial composition. In particular, it was found that the gut microbial composition was different between healthy individuals and patients with NAFLD (14, 15). Furthermore, mouse studies suggest that changes in the gut microbial community may impact the development of hepatic steatosis (16), inflammation (17, 18), and fibrosis (18).
To further elaborate the concept that the gut microbiota may affect the development of NAFLD, we studied the influence of modulation of the gut microbiota on NAFLD. To that end, in a mouse model of NAFLD, we stimulated the gut bacteria by feeding mice the highly fermentable dietary fiber, guar gum (GG), using the poorly fermentable dietary fiber, resistant starch (RS), as control (CTRL). Inasmuch as GG escapes digestion in the small intestine and is thought to act exclusively via bacterial fermentation and concomitant alterations in gut microbiota, feeding GG is an attractive strategy to study the role of the gut microbes in NAFLD. Additionally, in the same mouse model of NAFLD, we studied the effect on NAFLD of suppressing the gut bacteria using a mixture of antibiotics.
Overall, our data indicate that the gut microbiota have a marked impact on NAFLD. Specifically, specific modulation of the gut microbiota by feeding GG worsened features of NAFLD, whereas suppression of the gut bacteria using oral antibiotics protected against NAFLD, possibly via changes in the portal delivery of bile acids.
MATERIALS AND METHODS
Animals and diet
Animal studies were performed using pure-bred male C57Bl/6 mice. Mice were individually housed in temperature- and humidity-controlled specific pathogen-free conditions. Mice had ad libitum access to food and water. In study 1, 11-week-old mice received a low-fat diet or a high-fat/high-cholesterol/high-fructose diet (HFCFD) for 18 weeks, providing 10% or 45% energy as triglycerides (formula D12450B or D12451 from Research Diets, Inc., manufactured by Research Diet Services, Wijk bij Duurstede, The Netherlands). The fat source of this HFCFD was replaced by safflower oil and supplemented with 1% cholesterol (Dishman, Veenendaal, The Netherlands). Mice receiving the HFCFD were provided with 20% fructose water (w/v) to promote development of NAFLD (19). The fiber-enriched diets were identical to the HFCFD (CTRL) except that corn starch and part of the maltodextrin 10 were replaced by 10% (w/w) RS or GG. The composition of the diets is shown in supplemental Table S1. RS, a retrograded tapioca starch classified as RS type 3 (brand name C*Actistar 11700) and GG (brand name Viscogum) were obtained from Cargill R&D Centre Europe (Vilvoorde, Belgium). Bodyweight and food intake were assessed weekly. One mouse fed the GG-enriched diet died during the study for reasons unrelated to the intervention.
In study 2, 10-week-old mice received a HFCFD (CTRL) for 18 weeks, providing 45% energy as triglycerides (formula 58V8 manufactured by TestDiet, St. Louis, MO). This diet was supplemented with 1% cholesterol (Dishman). The mice received 20% fructose (w/v) in the drinking water. The trimethylamine N-oxide (TMAO)-enriched diet was identical to the CTRL diet except that 0.296% of sucrose (w/w) was replaced by 0.296% TMAO-dihydrate (w/w) (Sigma, Zwijndrecht, The Netherlands) to obtain a final concentration of 0.2% TMAO (w/w). Bodyweight and food intake were assessed weekly. One mouse fed the TMAO-enriched diet died during the study for reasons unrelated to the intervention. One mouse in the CTRL group and one mouse fed the TMAO-enriched diet failed to thrive and were excluded from further analysis.
In study 3, 4-month-old male mice received either chow (CTRL) or chow supplemented with 0.5% (w/w) taurocholic acid (Calbiochem) for 7 days (20). Bodyweight and food intake were assessed daily.
In study 4, 10-week-old mice received a HFCFD (CTRL) for 18 weeks, providing 45% energy as triglycerides (formula 58V8 manufactured by TestDiet). The diet was supplemented with 1% cholesterol (Dishman). The mice received 20% fructose (w/v) in the drinking water. Mice that were given antibiotics received the same CTRL diet with the addition of 1 g/l ampicillin, 1 g/l neomycin sulfate, 1 g/l metronidazole, and 0.5 g/l vancomycin in the drinking water for 22 weeks. This antibiotic cocktail has previously been shown to deplete all detectable commensal bacteria (21). Bodyweight and food intake were assessed weekly. One mouse in the CTRL group and one mouse in the antibiotics group failed to thrive and were excluded from further analysis.
At the end of each study, mice were anesthetized using isoflurane and blood was collected by orbital puncture. Mice were euthanized via cervical dislocation after which tissues were excised and weighed and intestinal content was sampled. The mice were not fasted prior to euthanasia (e.g., ad libitum fed state). The animal experiments were approved by the local animal ethics committee of Wageningen University.
Intraperitoneal glucose tolerance test
In study 1, mice were fasted for 5 h prior to the intraperitoneal glucose tolerance test. During this period, fructose water was replaced by tap water. Blood samples were collected from the tail vein immediately before (t = 0 min) and at selected time points after intraperitoneal injection with glucose (0.8 g/kg bodyweight). Glucose was measured using Accu-chek Compact. Plasma concentrations of insulin were quantified after 5 h fasting according to manufacturer’s instructions (Crystal Chem, Downers Grove, IL).
RNA isolation and quantitative PCR
Total RNA was extracted from epididymal white adipose tissue, liver, and scrapings of the distal small intestine using TRIzol reagent (Life Technologies, Bleiswijk, The Netherlands). Subsequently, 500 ng RNA was used to synthesize cDNA using iScript cDNA synthesis kit (Bio-Rad Laboratories, Veenendaal, The Netherlands). Changes in gene expression were determined by real-time PCR on a CFX384 real-time PCR detection system (Bio-Rad Laboratories) by using SensiMix (Bioline; GC Biotech, Alphen aan den Rijn, The Netherlands). The housekeeping gene, 36b4 or β-actin, was used for normalization. Sequences of the used primers are listed in supplemental Table S2.
Histology/immunohistochemistry
H&E staining of sections was performed using standard protocols.
Paraffin-embedded liver sections (5 μm) were stained for collagen using fast green FCF/Sirius red F3B. Staining of neutral lipids was performed on frozen liver sections using Oil Red O according to standard protocols.
Visualization of hepatic stellate cells was performed on paraffin-embedded liver sections with an antibody against α smooth muscle actin (αSMA) (M0851; Dako, Heverlee, Belgium). To detect macrophages, 5 μm frozen liver sections were stained for Cd68 Kupffer cells (Cd68 marker, FA11) as described previously (22).
Plasma/serum measurements
In studies 1 and 3, blood was collected in EDTA-coated tubes and centrifuged for 15 min at 1,000 g to obtain plasma. In studies 2 and 4, blood was collected, allowed to clot for 45 min, and centrifuged for 15 min at 1,000 g to obtain serum. The mice were not fasted prior to blood collection. Plasma and serum alanine aminotransferase (ALT) activity was measured with a kit from Abcam (Cambridge, UK). The commercially available Limulus Amebocyte Lysate assay (Lonza, Walkersville, MD) was used to quantify plasma and portal serum endotoxin levels. Plasma concentration of total bile acids was determined using a colorimetric assay kit (Diazyme Laboratories, Poway, CA). Free and conjugated bile acid subspecies were quantified by LC-MS/MS using a SHIMADZU LC system (SHIMADZU, Kyoto, Japan) and tandem AB SCIEX API-3,200 triple quadrupole mass spectrometers (AB SCIEX, Framingham, MA), as previously described (23). Trimethylamine (TMA) and TMAO plasma levels and TMAO serum levels were quantified by stable isotope dilution LC-MS/MS, as previously described (24, 25).
Fecal measurements
In study 1, during the sixth week of dietary intervention, feces were collected over a period of 48 h. Total lipids and free fatty acids were measured in the fecal samples as mentioned by Govers and Van der Meet (26). Briefly, 100 mg of fecal samples were weighed, dried, and acidified using HCl. Lipids were then extracted using petroleum and diethyl ether. The ether fraction was collected and evaporated and the total lipids were weighed.
Free and conjugated bile acid subspecies in the feces, collected at the end of the dietary intervention in studies 1 and 4, were determined by capillary gas chromatography (Agilent 6890; Amstelveen, The Netherlands) as described previously (27).
Microarray analysis
Microarray analysis was performed on liver samples from eight mice of the CTRL group and eight mice of the GG group. RNA was isolated as described above and subsequently purified using the RNeasy Micro kit from Qiagen (Venlo, The Netherlands). RNA integrity was verified with RNA 6000 Nano chips on an Agilent 2100 bioanalyzer (Agilent Technologies, Amsterdam, The Netherlands). Purified RNA (100 ng) was labeled with the Ambion WT expression kit (Invitrogen, Carlsbad, CA) and hybridized to an Affymetrix Mouse Gene 1.1 ST array plate (Affymetrix, Santa Clara, CA). Hybridization, washing, and scanning were carried out on an Affymetrix GeneTitan platform according to the manufacturer’s instructions. Arrays were normalized using the Robust Multi-array Average method (28, 29). Probe sets were defined according to Dai et al. (30). In this method, probes are assigned to Entrez IDs as a unique gene identifier. The P values were calculated using an intensity-based moderated t-statistic (31). The q value was calculated as the measure of significance for the false discovery rate (32). To identify the pathways most significantly altered in the livers of the GG group compared with the CTRL group, Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) was performed. Input criteria were a relative fold change equal to or above 1.3 and a q value equal to or below 0.01. Array data have been submitted to the Gene Expression Omnibus under accession number GSE76087.
A publicly available dataset (GSE34730) was downloaded from Gene Expression Omnibus and further processed as described above to obtain individual gene expression data. Ingenuity Pathway Analysis was performed on genes induced by deoxycholic acid by at least 2-fold.
Western blot
Liver homogenates were prepared by lysing liver tissue in ice-cold Pierce IP lysis buffer (ThermoScientific, Rockford, IL) containing complete protease inhibitor and PhosSTOP phosphatase inhibitor (both from Roche, Mannheim, Germany). Equal amounts of protein were loaded on a Mini Protean TGX gel, 4–15%, and subsequently transferred to a PVDF membrane (both from Bio-Rad Laboratories). The primary antibodies for tissue inhibitor of metalloproteinase (TIMP)1 (Abcam, Cambridge, UK) and HSP90 (Cell Signaling) were incubated at 4°C overnight followed by incubation with the appropriate secondary peroxidase-conjugated antibody (Sigma). Protein bands were visualized using an enhanced chemiluminescent substrate (Bio-Rad Laboratories).
Liver measurements
For liver triglyceride measurement, livers were homogenized in a buffer containing 10 mM Tris, 2 mM EDTA, and 250 mM sucrose at pH 7.5 in a Tissue Lyser II (Qiagen, Hilden, Germany) to obtain 2% homogenates. Triglycerides were subsequently quantified using a Triglyceride LiquiColormono kit from HUMAN Diagnostics (Wiesbaden, Germany). The 4-hydroxyproline content was determined spectrophotometrically in liver hydrolysates, as described previously (33).
For hepatic myeloperoxidase activity, liver homogenates were prepared and myeloperoxidase peroxidase activity was measured according to manufacturer’s instructions (Abcam).
For the quantification of liver total bile acid content, livers were homogenized in 75% ethanol, incubated at 50°C and centrifuged. After the collection of the supernatant, the concentration of total bile acids was determined using a colorimetric assay kit (Diazyme Laboratories).
FACS analysis
Epididymal white adipose tissue was isolated, minced and digested with 1.5 mg/ml collagenase type 2 (Sigma) in DMEM containing 0.5% fatty acid-free BSA for 45 min at 37°C by shaking. Cell suspensions were filtered through a 100 μM filter and centrifuged at 200 g for 10 min. Floating mature adipocytes were discarded and the stromal vascular cell pellet was resuspended in erythrocyte lysis buffer for 5 min. Cells were washed twice with FACS buffer (PBS + 1% BSA) and incubated with fluorescently labeled antibodies including CD45-ECD (Beckman Coulter, Fullerton, CA), F4/80-FITC and Cd11b-PE (Biolegend, San Diego, CA). Samples were analyzed using a FC500 flow cytometer (Beckman Coulter).
DNA extraction
In study 1, fecal samples derived from the second part of the colon were suspended in 10 mM Tris, 1 mM EDTA, 0.5% SDS, and 0.2 mg/ml proteinase K (ThermoScientific). After addition of 0.1–0.25 mm and 4 mm glass beads, buffered phenol (Invitrogen) was added and cells were lysed by mechanical disruption using a bead beater (MP Biomedicals, Santa Ana, CA) for 3 min. DNA was subsequently extracted using phenol:chloroform:isoamylalcohol (25:24:1) (Invitrogen), precipitated with isopropanol, and washed with 70% ethanol.
In study 4, fecal samples (∼15–60 mg) derived from the second part of the colon were suspended in 500 μl S.T.A.R. buffer (Roche). After addition of 0.1 mm zirconia and 2.5 mm glass beads (BioSpec, Bartlesville, OK), cells were lysed by mechanical disruption using a bead beater (MP Biomedicals) for 3 × 1 min. DNA was subsequently extracted and purified using the Maxwell 16 system (Promega). In brief, homogenates (250 μl) were transferred to a prefilled reagent cartridge (Maxwell® 16 Tissue LEV Total RNA purification kit, custom-made; Promega). Sixteen samples were processed at the same time. After 30 min, the purification process was completed and DNA was eluted in 50 μl of water (nuclease free) (Promega).
The 16S rRNA gene sequencing
For 16S rRNA gene sequencing DNA samples were sent to the Broad Institute of Massachusetts Institute of Technology and Harvard (Cambridge, MA). The microbial 16S rRNA gene was amplified targeting the hyper-variable region V4 using forward primer 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The cycling conditions consisted of an initial denaturation of 94°C for 3 min, followed by 25 cycles of denaturation at 94°C for 45 s, annealing at 50°C for 60 s, extension at 72°C for 5 min, and a final extension at 72°C for 10 min. Sequencing was performed using the Illumina MiSeq platform generating paired-end reads of 175 bp in length in each direction. Overlapping paired-end reads were subsequently aligned. The details of this protocol are as previously described (34).
Raw sequence data quality was assessed using FastQC, version: 0.11.2 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads quality was checked with Sickle, version: 1.33 (https://github.com/najoshi/sickle) and low quality reads were removed. For visualizing the taxonomic composition of the fecal microbiota and further β diversity analysis, QIIME, version: 1.9.0 was used (35). In brief, closed reference OTU picking with 97% sequence similarity against GreenGenes 13.8 reference database was done. Jackknifed β-diversity of unweighted UniFrac distances with 10 jackknife replicates was measured at rarefaction depth of 22,000 reads per sample. For statistical significance, biological relevance, and visualization, we used the linear discriminant analysis (LDA) effect size (LEfSe) method (https://bitbucket.org/biobakery/biobakery/wiki/lefse) (36).
Bacterial 16S rRNA gene and fungal ITS1-5.8S-ITS2 region quantification
In study 4, real-time PCR was used to quantify the 16S rRNA gene and ITS1-5.8S-ITS2 region in equal amounts of extracted DNA on a CFX384 real-time PCR detection system (Bio-Rad Laboratories) by using SensiMix (Bioline; GC Biotechnology). The 16S rRNA gene was amplified using the forward primer 1369F (5′-CGGTGAATACGTTCYCGG-3′) (37) and the reverse primer 1492R (5′-GGWTACCTTGTTACGACTT-3′) (38). The cycling conditions consisted of an initial denaturation of 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s The ITS1-5.8S-ITS2 region was amplified using universal fungal primers, V9D (5′-TTAAGTCCCTGCCCTTTGTA-3′) and LS266 (5′-GCATTCCCAAACAACTCGACTC-3′), encompassing highly conserved regions encoding fungal rDNA (39). The cycling conditions consisted of an initial denaturation of 94°C for 10 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 30 s, and a final extension at 72°C for 10 min.
1H NMR spectroscopy
Contents of the cecum and first part of the colon were collected, mixed with phosphate buffer (pH 8) containing 10% deuterium oxide and stored at −20°C. For short-chain fatty acid (SCFA), ethanol and choline quantification samples were thawed, mixed, and centrifuged at 21,000 g for 5 min. Supernatant was collected and diluted in phosphate buffer containing 2 mM maleic acid as standard. Subsequently, a 200 μl sample was transferred to 3 mm NMR tubes (Bruker Match System) and measured at 300 K in an Avance III NMR spectrometer operated at 600.13 MHs, as described previously (40).
Statistical analysis
Data are presented as mean ± SEM. Statistically significant differences were determined with one-way ANOVA followed by Tukey’s post hoc multiple comparison test. Comparisons between two groups were made using two-tailed Student’s t-test. P < 0.05 was considered as statistically significant. SPSS software (version 21; SPSS Inc., Chicago, IL) was used for statistical analysis.
RESULTS
HFCFD as model for NAFLD
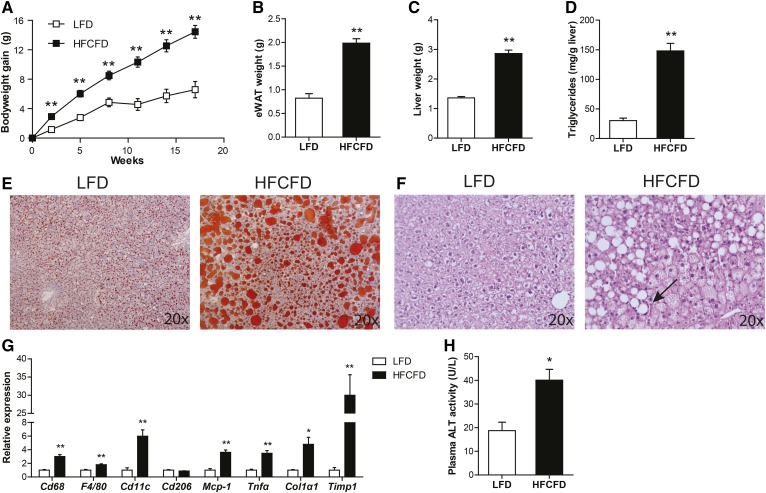
Feeding mice a diet rich in fat, cholesterol, and/or fructose is a commonly used strategy to induce NAFLD (41–43). To verify the suitability of the HFCFD as a model for NAFLD, we tested the effect of HFCFD on various relevant parameters in comparison with a commonly used low-fat diet. Weight gain, adipose tissue weight, and liver weight were significantly higher in the mice fed HFCFD as compared with mice fed the low-fat diet (Fig. 1A–C). Hepatic lipid storage was markedly higher in mice fed HFCFD, as shown by a 5-fold increase in liver triglycerides (Fig. 1D) and by Oil Red O staining (Fig. 1E). H&E confirmed the presence of microvesicular and macrovasicular steatosis, and showed mild infiltration of immune cells (Fig. 1F). Hepatic expression of macrophage markers, Cd68, F4/80, and Cd11c, and pro-inflammatory cytokines, Mcp-1 and Tnfα, were significantly elevated in mice fed HFCFD, as was the expression of fibrosis markers, collagen type I α1, and Timp1 (Fig. 1G). Finally, plasma ALT levels were doubled in mice fed HFCFD (Fig. 1H). Taken together, these data show that feeding HFCFD represents a suitable model to study NAFLD in mice.
Fig. 1.
Effect of HFCFD on obesity and features of NAFLD. A: Bodyweight gain in C57Bl/6 mice fed a low-fat diet (LFD) or a HFCFD. Weight of epididymal white adipose tissue (eWAT) (B) and liver (C) after 18 weeks of dietary intervention. D: Quantitative analysis of liver triglyceride levels. Oil Red O staining (E) and H&E staining (F) of representative liver sections. Arrow denotes inflammatory infiltrate. G: Relative expression of inflammation and fibrosis-related genes in the liver. Gene expression levels of mice fed a low-fat diet were set at one. H: Activity of ALT in plasma. Data are presented as mean ± SEM. Asterisks indicate significantly different compared with low-fat diet. *P < 0.05, **P ≤ 0.001.
Dietary GG alters colonic microbial composition
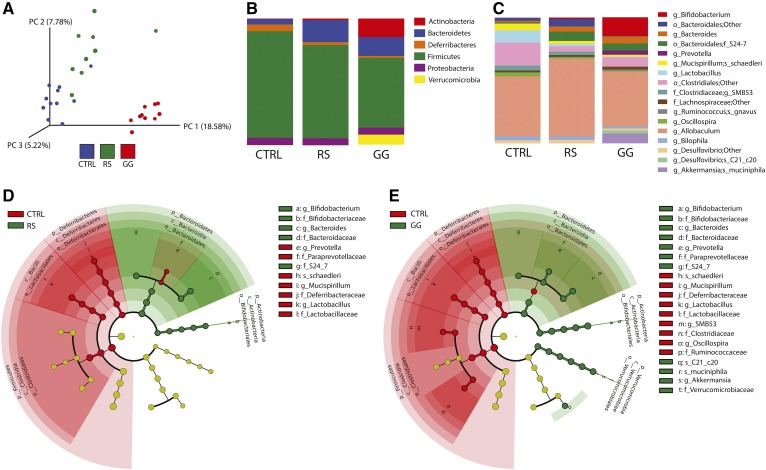
To evaluate the potential impact of alterations in the gut microbial community on the development of NAFLD, we fed mice the highly fermentable dietary fiber, GG, on a background of HFCFD for 18 weeks. Mice fed the poorly fermentable dietary fiber, RS, and mice that were not fed any dietary fiber (CTRL) served as negative CTRLs. To assess the effect of GG on colonic microbial composition and the relative abundance of specific gut microbiota taxa, 16S rRNA gene sequencing was performed on fecal samples collected at the end of the dietary intervention. Clustering of 16S rRNA gene sequences by unweighted UniFrac distances per mouse revealed a sharp clustering of microbiome sequence data of the GG mice, indicating that the colonic microbial community was distinct between CTRL and GG mice. In contrast, the microbiota of the RS mice more closely resembled that of the CTRL mice (Fig. 2A). Analysis of the microbiota at various taxonomic levels indicated differences in microbial composition depending on fiber type. In comparison with CTRL, both types of dietary fibers significantly reduced the relative abundance of Deferribacteres and Firmicutes, and increased the relative abundance of Bacteroidetes. The abundance of Actinobacteria was particularly increased in the GG mice. With a relative proportion of 8.23% of all phyla, the phylum Verrucomicrobia was overrepresented in the GG mice, whereas it was almost completely absent in CTRL and RS mice (Fig. 2B, supplemental Table S3).
Fig. 2.
GG alters the colonic microbial composition. A: Principle coordinates analysis plot of unweighted UniFrac distances indicating clustering 16S rRNA gene sequences. Each circle represents an individual mouse. B: Mean relative abundance of the predominant phyla (>0.5% in at least one of the three diet groups). C: Mean relative abundance of bacteria reported to the lowest identifiable level. Taxonomic level is indicated by letter preceding the underscore: o, order; f, family; g, genus; s, species. D: Cladogram representing significant enrichment of taxa in CTRL (red) or RS (green) colonic microbiota. E: Cladogram representing significant enrichment of taxa in CTRL (red) or GG (green) colonic microbiota. The central point in the cladogram represents the domain bacteria and each ring represents the next lower taxonomic level (phylum to genus).
The differences in microbial community between the RS and GG groups became more apparent at lower taxonomic levels. Whereas GG increased the abundance of the genera Bifidobacterium and Prevotella, RS had relatively small effects on these genera. The decreased abundance of the phylum Firmicutes for both dietary fibers can be mainly explained by a decrease within the genus Lactobacillus. GG also markedly suppressed SMB53 and Oscillospira and increased the abundance of the species Desulfovibrio C21_c20 (Fig. 2C–E, supplemental Table S3). Overall, these data indicate that GG feeding markedly altered the bacterial composition in the colon.
Modulation of the gut microbiota by GG is associated with protection against diet-induced obesity and improved glucose tolerance
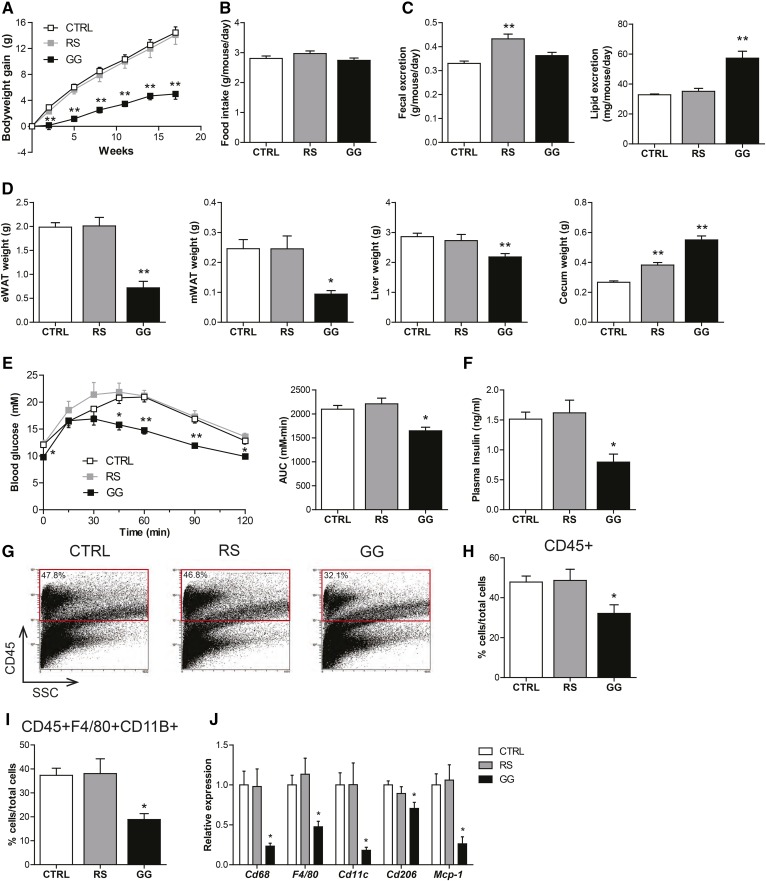
Whereas RS did not influence bodyweight gain, GG markedly attenuated bodyweight gain as compared with CTRL (Fig. 3A). The lower bodyweight could not be attributed to reduced food intake (Fig. 3B) or an increased fecal output (Fig. 3C). Fecal lipid excretion was significantly higher in the GG mice as compared with the CTRL mice (Fig. 3C), suggesting that the lower bodyweight in the GG mice might be explained by decreased lipid absorption. Along with the lower bodyweight, weights of epididymal and mesenteric fat pads and liver were significantly lower in the GG group as compared with the CTRL group, while the weight of the cecum was higher in both RS and GG groups (Fig. 3D).
Fig. 3.
Stimulation of the gut microbiota by GG is associated with protection against the development of diet-induced obesity, improved glucose tolerance, and decreased adipose tissue inflammation. A: Changes in bodyweight in C57Bl/6 mice fed a HFCFD (CTRL) or a HFCFD supplemented with RS or GG. B: Mean food intake per day during the dietary intervention. C: Mean fecal and lipid excretion per mouse per day in the sixth week of dietary intervention. D: Weight of epididymal white adipose tissue (eWAT), mesenteric white adipose tissue (mWAT), liver, and cecum after 18 weeks of dietary intervention. E: Plasma glucose levels of CTRL, RS, and GG mice following an intraperitoneal glucose tolerance test (GTT) and areas under the curve (AUC) after 17 weeks of dietary intervention. F: Plasma insulin concentration after 5 h of fasting. G: Representative flow cytometry plots and (H) corresponding quantitative analysis of infiltrated leukocytes (CD45+) in epididymal white adipose tissue (eWAT). I: Quantitative analysis of macrophages (F4/80+CD11B+). J: Relative expression of inflammatory genes in eWAT. Gene expression levels in CTRL mice were set at one. Data are presented as mean ± SEM. Asterisks indicate significantly different compared with CTRL. *P < 0.05, **P < 0.001.
In line with the suppression of bodyweight gain, GG improved whole body glucose tolerance (Fig. 3E), reduced fasting plasma insulin levels (Fig. 3F), and attenuated immune cell infiltration in adipose tissue. Specifically, flow cytometry analysis demonstrated that the abundance of leukocytes (CD45+) and macrophages (CD45+F4/80+CD11B+) was significantly lower in the epididymal fat depot of the GG mice as compared with the CTRL mice, whereas it was not different in the RS mice (Fig. 3G–I). The decreased macrophage abundance was corroborated by decreased expression of the chemoattractant protein, Mcp-1, and macrophage markers, F4/80 and Cd68. Cd11c and Cd206 mRNA were also decreased in the GG group, suggesting lower presence of pro-inflammatory M1 macrophages and anti-inflammatory M2 macrophages, respectively (Fig. 3J). Taken together, GG suppressed bodyweight gain, which was accompanied by decreased adipose tissue inflammation and improved glucose tolerance.
Modulation of the gut microbiota by GG is associated with reduced hepatic steatosis, but enhanced hepatic inflammation
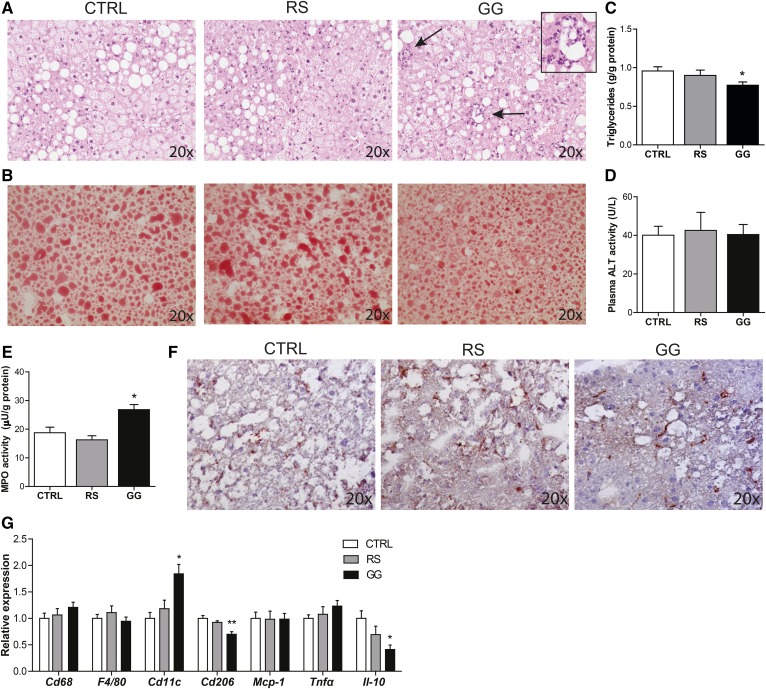
In accordance with a lower fat mass in the GG mice, hepatic lipid accumulation was significantly lower in mice fed GG, as revealed by H&E and Oil Red O stainings and quantitation of liver triglycerides (Fig. 4A–C). Plasma ALT activity, however, was not different between the three groups (Fig. 4D). Interestingly, despite the reduction in liver triglycerides, H&E staining showed more pronounced inflammation in the livers of GG mice, as revealed by the presence of inflammatory infiltrates (Fig. 4A). Consistent with these data, elevated myeloperoxidase activity (Fig. 4E) indicated a higher number of neutrophils in the livers of the GG mice. In contrast, the total macrophage abundance assessed by Cd68 staining and Cd68 and F4/80 expression was not different between the GG and CTRL mice (Fig. 4F, G). Intriguingly, the higher expression of Cd11c and the lower expression of Cd206 and the anti-inflammatory cytokine Il-10 suggested that the macrophages present in the liver of the mice fed GG had a more pro-inflammatory phenotype (Fig. 4G). To further elucidate the inflammatory status of the liver of the GG mice, whole genome expression profiling was performed. It was found that expression of 3,722 genes was significantly different between the GG mice compared with the CTRL mice (P < 0.01) (supplemental Fig. S1A). Numerous immune-related pathways, including interleukin (IL)-8 signaling (P = 1.05 × 10−6) and complement system activation (P = 7.76 × 10−7), were altered in the livers of the GG mice, as determined by Ingenuity Pathway Analysis (supplemental Fig. S1B). Together, these data demonstrate that, although feeding GG suppressed diet-induced obesity, it stimulated specific inflammatory cells and pathways in the liver, whereas RS had no effect.
Fig. 4.
Stimulation of the gut microbiota by GG is associated with reduced hepatic steatosis, but enhanced hepatic inflammation. A: H&E staining of representative liver sections. Arrows denote inflammatory infiltrate and inset a higher magnification of the inflammatory infiltrate as observed in the livers of the GG mice. B: Representative pictures of Oil Red O staining of liver sections. C: Quantitative analysis of liver triglyceride levels. D: Activity of ALT in plasma. E: Liver myeloperoxidase (MPO) activity representing hepatic neutrophil content. F: Representative pictures of liver sections stained for the macrophage marker Cd68. G: Relative expression of inflammatory genes in the liver. Gene expression levels in CTRL mice were set at one. Data are presented as mean ± SEM. Asterisks indicate significantly different compared with CTRL. *P < 0.05, **P < 0.001.
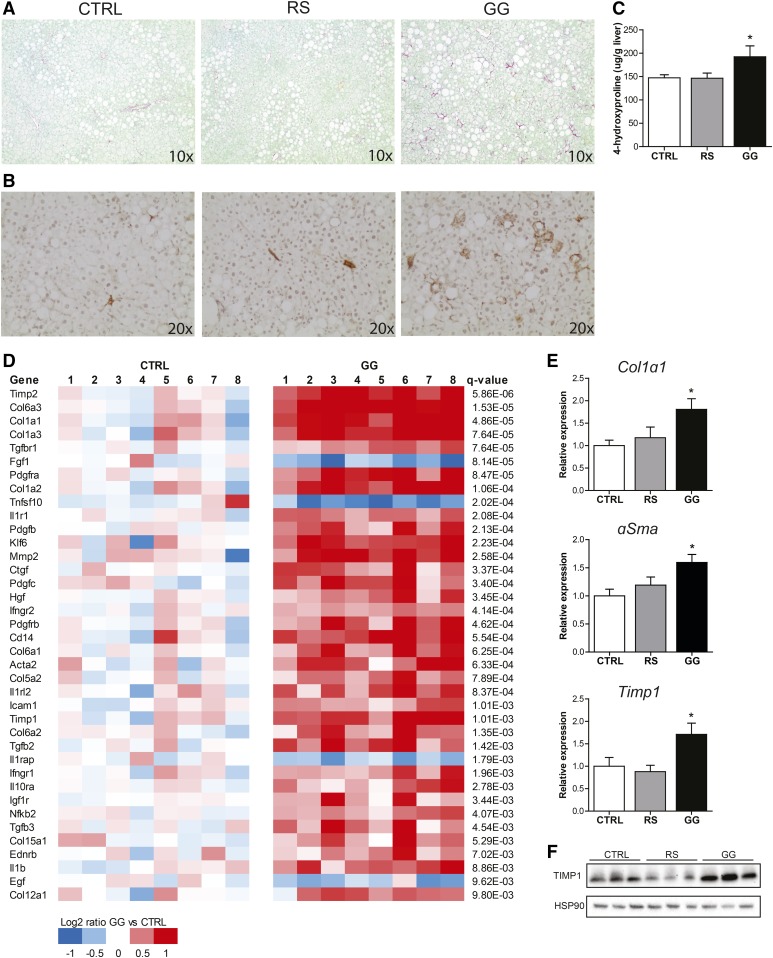
Modulation of the gut microbiota by GG is associated with enhanced hepatic fibrosis
Strikingly, further study revealed that the livers of the mice fed GG exhibited pronounced features of hepatic fibrosis. Sirius red staining showed enhanced collagen deposition in the GG mice as compared with CTRL and RS mice (Fig. 5A). Likewise, hepatic stellate cell activation, as visualized by αSMA immunostaining and 4-hydroxyproline content, were also increased in the GG mice (Fig. 5B, C). Expression profiling followed by Ingenuity Pathway Analysis pointed to hepatic fibrosis/hepatic stellate cell activation as the most significantly regulated pathway (P = 7.94 × 10−11), which was illustrated by the differential expression of numerous genes involved in hepatic fibrosis and hepatic stellate cell activation (Fig. 5D). Genes highly induced in the livers of GG mice included various collagens and genes involved in the regulation of extracellular matrix turnover (Timp2) and TGFβ signaling (Tgfbr1). Elevated expression of fibrosis-related genes was corroborated by quantitative (q)PCR (Fig. 5E). Finally, TIMP1 protein levels were elevated in the livers of the GG mice (Fig. 5F). These data indicate that feeding GG not only increased hepatic inflammation, but also promoted more advanced features of NASH, i.e., liver fibrosis.
Fig. 5.
Stimulation of the gut microbiota by GG is associated with enhanced hepatic fibrogenesis. Representative pictures of liver sections stained for collagen using fast green FCF/Sirius red F3B (A) and hepatic stellate cell activation using an antibody against αSMA (B). C: The 4-hydroxyproline content in the liver. D: Heat map showing significant changes in expression of genes involved in hepatic fibrosis/hepatic stellate cell activation in the livers of CTRL and GG mice. The Log2 expression signals of the CTRL group were arbitrarily set at zero. E: Relative expression of fibrosis-related genes in the liver. Gene expression levels in CTRL mice were set at one. Data are presented as mean ± SEM. Asterisks indicate significantly different compared with CTRL. *P < 0.05. F: TIMP1 protein expression measured by Western blotting. HSP90 was used as loading control.
Modulation of the gut microbiota by GG is associated with altered levels of gut-derived metabolites
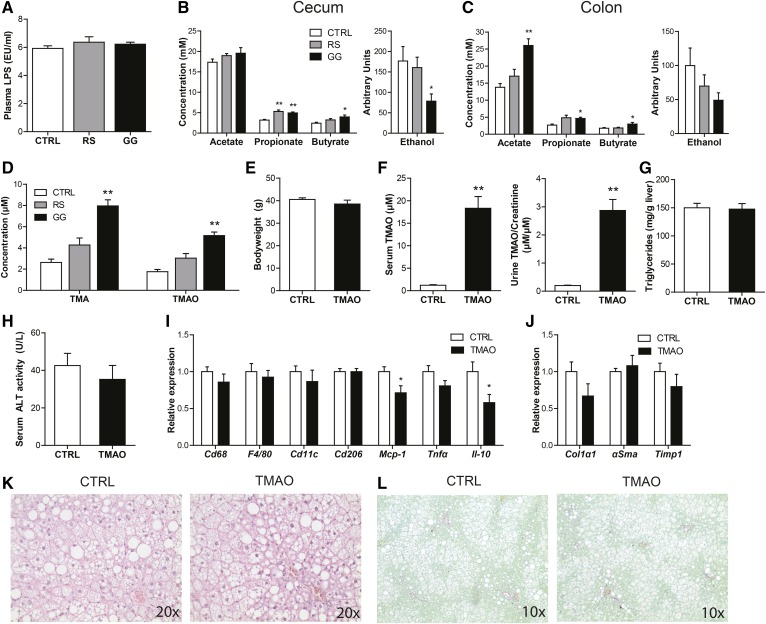
To explore the possible mechanisms linking changes in gut microbiota composition to the observed liver pathology, we focused our attention on several microbial compounds that have been proposed to mediate the effects of the gut bacteria on host metabolism, including the pro-inflammatory lipopolysaccharide (LPS) and ethanol, and the anti-inflammatory SCFAs (44). Plasma levels of LPS were comparable between the GG mice and CTRL mice (Fig. 6A), while ethanol levels in the cecum and colon were decreased in the GG group (Fig. 6B, C). With respect to the SCFA, in the cecum, the concentration of propionate was increased in both dietary fiber groups, whereas the butyrate concentration was only elevated in the GG group (Fig. 6B). In the colon, GG, but not RS, significantly elevated acetate, propionate, and butyrate concentrations, as compared with CTRL (Fig. 6C). In short, the observed changes in NAFLD phenotype in the GG mice are unlikely to be mediated by LPS, ethanol, or SCFAs.
Fig. 6.
GG-induced elevation of TMAO does not contribute to enhanced NALFD development. A: Plasma LPS levels. Cecal (B) and colonic (C) SCFA concentration and ethanol abundance. D: Plasma TMA and TMAO levels. Mice were fed the HFCFD for 18 weeks with or without supplementation with TMAO. E: Final bodyweight. F: Serum and urinary TMAO levels. Urinary TMAO levels are corrected for creatinine levels to adjust for urinary dilution. G: Quantitative analysis of liver triglyceride levels. H: Activity of ALT in serum. Relative expression of inflammation (I) and fibrosis-related (J) genes in the liver. Gene expression levels in CTRL mice were set at one. Data are presented as mean ± SEM. Asterisks indicate significantly different compared with CTRL. *P < 0.05, **P < 0.001. K: H&E staining of representative liver sections. L: Representative pictures of liver sections stained for collagen using fast green FCF/Sirius red F3B.
Interestingly, plasma levels of the gut-derived metabolites, TMA, a product of microbial conversion of choline, and TMAO, which is synthesized from TMA in the liver, were both significantly increased by GG, while RS had no effect (Fig. 6D). Previously, TMAO was causally implicated in atherosclerosis and kidney fibrosis (9, 45, 46).
Accordingly, to investigate whether the elevated plasma TMAO levels may contribute to the NAFLD phenotype, mice were fed the HFCFD diet with or without 0.2% TMAO (w/w) for 18 weeks. Eighteen weeks of TMAO feeding did not affect final bodyweight (Fig. 6E). Although serum and urinary TMAO levels were about 15-fold higher in the mice fed the TMAO-enriched diet (Fig. 6F), provision of TMAO did not affect hepatic steatosis, inflammation, or fibrosis, as indicated by measurement of liver triglycerides (Fig. 6G), serum ALT activity (Fig. 6H), the expression of inflammation and fibrosis-related genes (Fig. 6I, J), H&E staining (Fig. 6K), and Sirius red staining (Fig. 6L).
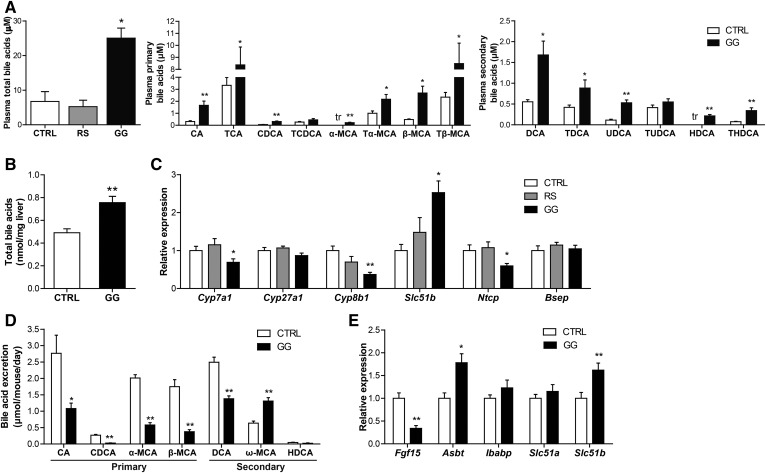
Bile acids are also potential candidates for linking changes in gut bacteria to NAFLD. Strikingly, total plasma bile acid levels were nearly 4-fold higher in the GG mice than in the CTRL mice, whereas they were not different in the RS mice (Fig. 7A). Specifically, plasma levels of various primary bile acids, including cholic acid, taurocholic acid, muricholic acids, and tauromuricholic acids, as well as the secondary bile acids, deoxycholic acid, taurodeoxycholic acid, ursodeoxycholic acid, hyodeoxycholic acid, and taurohyodeoxycholic acid, were significantly higher in the GG mice as compared with CTRL mice (Fig. 7A). Moreover, total bile acid levels in the liver were also increased in the GG mice as compared with the CTRL mice (Fig. 7B). The higher plasma and hepatic bile acid levels were accompanied by elevated hepatic expression of the FXR target, Slc51b, and a compensatory downregulation of Cyp7a1 and Cyp8b1, enzymes involved in bile acid synthesis, and Ntcp (Slc10a1), a bile salt importer (Fig. 7C). No change was observed in the expression of the bile acid exporter, Bsep (Abcb11) (Fig. 7C). Interestingly, levels of bile acids in the feces were significantly lower in the GG mice (Fig. 7D), concomitant with reduced ileal expression of Fgf15 and elevated expression of the ileal bile acid transporters, Asbt (Slc10a2) and Slc51b (Fig. 7E). Together, these data suggest that GG enhanced bile acid absorption, leading to higher bile acid levels in the plasma and elevated bile acid content in the liver, which in turn triggered compensatory changes in the expression of genes involved in hepatic bile acid synthesis and uptake.
Fig. 7.
GG elevates plasma bile acid levels likely by promoting bile acid reabsorption. A: Total bile acid concentration in plasma of CTRL, RS, and GG mice and concentration of free and conjugated primary and secondary bile acid subspecies in the plasma of CTRL and GG mice. B: Total bile acid content in the liver of CTRL and GG mice. C: Relative expression of genes encoding enzymes of bile acid synthesis and bile acid transporters. Gene expression levels in CTRL mice were set at one. D: Mean excretion of free and conjugated bile acid subspecies per mouse per day in the CTRL and GG groups at the end of the dietary intervention. E: Relative expression of Fgf15 and bile acid transporters in the ileum. Gene expression levels in CTRL mice were set at one. Data are presented as mean ± SEM. Asterisks indicate significantly different compared with CTRL. *P < 0.05, **P < 0.001.
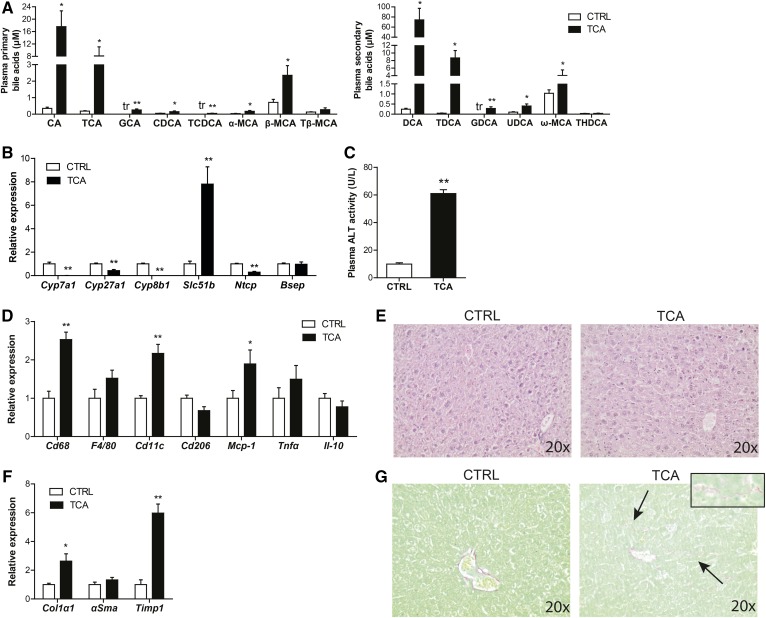
To examine whether the elevated plasma levels of bile acids may contribute to hepatic inflammation and fibrosis, we gave mice the bile acid, taurocholic acid, for 7 days by mixing it in their feed [0.5% (w/w)]. Provision of taurocholic acid raised plasma taurocholic acid levels by 40-fold as compared with CTRL (Fig. 8A). In addition, taurocholic acid also significantly increased the primary bile acids, cholic acid, glycocholic acid, chenodeoxycholic acid, taurochenodeoxycholic acid, and unconjugated muricholic acids, as well as the secondary bile acids, deoxycholic acid, taurodeoxycholic acid, glycodeoxycholic acid, ursodeoxycholic acid, and ω-muricholic acid (Fig. 8A). The higher plasma bile acid levels were associated with compensatory downregulation of Cyp7a1, Cyp27a1, Cyp8b1, and Ntcp and upregulation of Slc51b in liver (Fig. 8B). Importantly, provision of taurocholic acid markedly increased plasma ALT activity (Fig. 8C), upregulated the expression of inflammatory genes (Fig. 8D), and stimulated leukocyte infiltration (Fig. 8E). Moreover, taurocholic acid promoted hepatic fibrosis, as revealed by elevated expression of the fibrosis markers, collagen type I α1 and Timp1 (Fig. 8F), and enhanced collagen deposition (Fig. 8G). Analysis of a publicly available transcriptomics dataset (GSE34730) of livers of mice treated with cholic acid or deoxycholic acid for 2 weeks confirmed the marked stimulatory effect of oral administration of bile acids on genes implicated in hepatic inflammation and fibrosis. Specifically, Ingenuity Pathway Analysis showed the significant induction of numerous pathways related to immune cells and cytokine signaling, including IL-8 signaling and acute phase response signaling, as well as hepatic fibrosis/hepatic stellate activation. These effects were similarly observed in conventionalized and germ-free mice, indicating that the effects of oral cholic and deoxycholic acid on liver inflammation and fibrosis are not dependent on bacterial conversion (supplemental Fig. S2). Together, these data suggest that elevated plasma levels of bile acids can lead to hepatic inflammation and fibrosis in mice fed GG.
Fig. 8.
Bile acids promote hepatic injury. A: Concentration of free and conjugated bile acid subspecies in the plasma of CTRL and taurocholic acid-fed mice. The tr indicates that only trace levels of this bile acid could be measured. B: Relative expression of genes encoding enzymes of bile acid synthesis and bile acid transporters. Gene expression levels in CTRL mice were set at one. C: Activity of ALT in plasma. D: Relative expression of inflamation-related genes in the liver. Gene expression levels in CTRL mice were set at one. E: H&E staining of representative liver sections. F: Relative expression of fibrosis-related genes in the liver. Gene expression levels in CTRL mice were set at one. G: Representative pictures of liver sections stained for collagen using fast green FCF/Sirius red F3B. Inset is a higher magnification of the collagen deposition as observed in the livers of the taurocholic acid-fed mice. Data are presented as mean ± SEM. Asterisks indicate significantly different compared with CTRL. *P < 0.05, **P ≤ 0.001.
Suppression of the gut bacteria by oral antibiotics attenuates NAFLD development
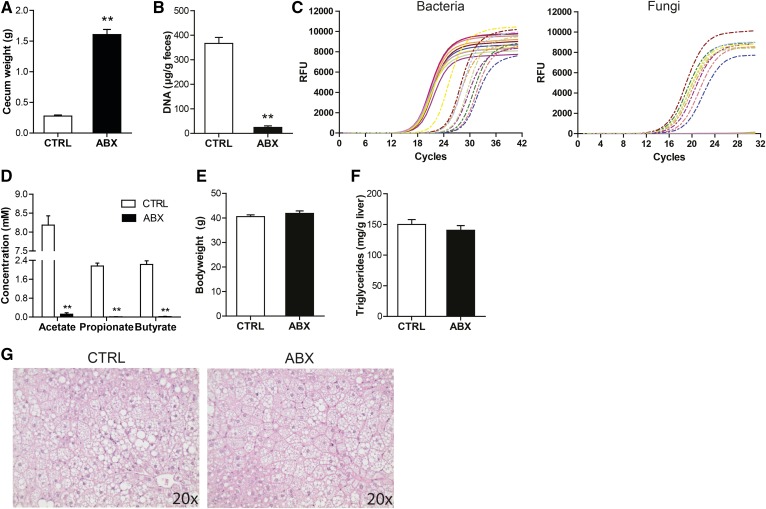
In order to study the direct impact of the gut microbiota on features of NAFLD, mice fed the HFCFD diet were given antibiotics via the drinking water for the entire duration of the dietary intervention. Oral administration of antibiotics led to a large increase in the weight of the cecum (Fig. 9A), and caused a massive decrease in fecal DNA levels (Fig. 9B) and fecal levels of the 16S rRNA gene (Fig. 9C). In addition, antibiotics dramatically reduced SCFA levels in the cecum (Fig. 9D), demonstrating the effective suppression of the bacteria in the colon. Interestingly, antibiotics triggered an outgrowth of fungi (Fig. 9C). At the end of the dietary intervention, final bodyweights were similar between the antibiotic-treated and CTRL mice (Fig. 9E). Antibiotic treatment did not affect hepatic steatosis development, as indicated by quantitative measurement of liver triglycerides (Fig. 9F) and H&E staining (Fig. 9G).
Fig. 9.
Suppression of the gut bacteria using antibiotics does not affect hepatic steatosis. A: Weight of the cecum. B: Quantitative analysis of fecal DNA levels. C: qPCR amplification plot for 16S rRNA gene and fungal ITS1-5.8S-ITS2 region in equal amounts of fecal DNA. Solid lines indicate CTRL mice and dashed lines indicate ABX mice. D: Cecal SCFA concentration. E: Final bodyweight of mice fed CTRL diet with or without antibiotic supplementation. F: Quantitative analysis of liver triglyceride levels. G: H&E staining of representative liver sections. Data are presented as mean ± SEM. Asterisks indicate significantly different compared with CTRL. **P ≤ 0.001.
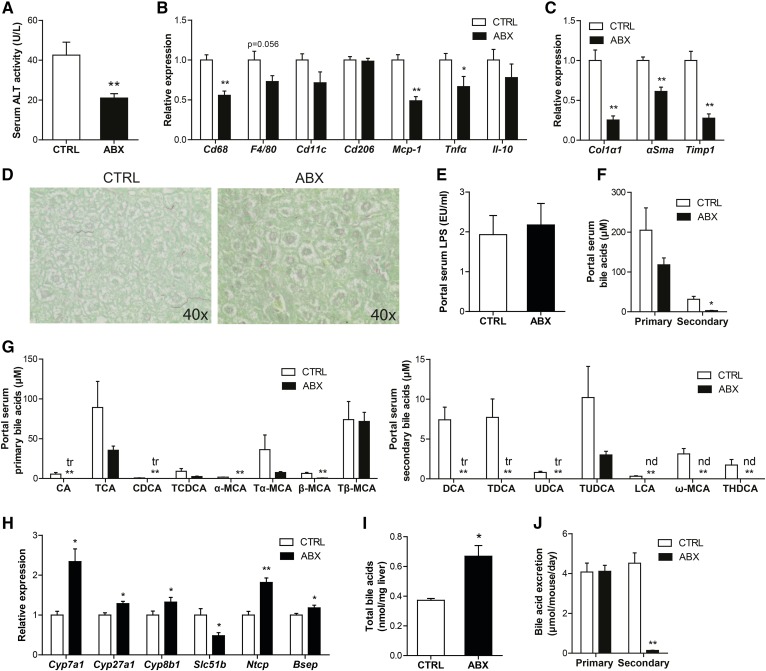
Strikingly, oral antibiotics caused a marked decrease in serum ALT activity (Fig. 10A), as well as a decrease in the expression of inflammatory markers in the liver (Fig. 10B). In addition, antibiotic treatment reduced hepatic fibrosis, as indicated by markedly lower expression of the fibrosis markers, collagen type I α1, αSMA, and Timp1 (Fig. 10C), and less collagen deposition (Fig. 10D). While LPS levels in portal blood were not different between CTRL and antibiotic-treated mice (Fig. 10E), antibiotic treatment led to a nonsignificant reduction in the portal concentration of primary bile acids and a highly significant 10-fold reduction in the concentration of secondary bile acids (Fig. 10F), hinting at a potential role of bile acids in the attenuation of features of NAFLD in the livers of the antibiotic-treated mice. Specifically, in the portal blood, the primary bile acids, cholic acid, chenodeoxycholic acid, and muricholic acids, and almost all secondary bile acids were reduced upon antibiotic treatment (Fig. 10G). The reduction in portal delivery of bile acids was accompanied by a significant reduction in hepatic expression of the FXR target, Slc51b, and a compensatory induction of genes involved in bile acid synthesis, including Cyp7a1, Cyp27a1, and Cyp8b1, as well as the bile acid importer, Ntcp, and exporter, Bsep (Fig. 10H). Surprisingly, total hepatic bile acid levels were increased in the antibiotic-treated mice (Fig. 10I). As expected, treatment with antibiotics caused a marked reduction in fecal excretion of secondary bile acids, whereas excretion of primary bile acids remained unchanged (Fig. 10J). Together, these data indicate that suppression of the gut bacteria attenuates features of NAFLD, possibly via reduced portal delivery of bile acids.
Fig. 10.
Antibiotics attenuate hepatic inflammation and fibrosis. A: Activity of ALT in serum. Relative expression of inflammation (B) and fibrosis-related (C) genes in the liver. Gene expression levels in CTRL mice were set at one. D: Representative pictures of liver sections stained for collagen using fast green FCF/Sirius red F3B. E: Portal serum LPS levels. F: Concentration of primary and secondary bile acids in portal serum. G: Concentration of free and conjugated primary and secondary bile acid subspecies in the portal serum of CTRL and GG mice. The tr indicates that only trace levels of this bile acid could be measured. The nd indicates that levels of this bile acid were below detectable limit. H: Relative expression of genes encoding enzymes of bile acid synthesis and bile acid transporters. Gene expression levels in CTRL mice were set at one. I: Total bile acid content in the liver. J: Mean excretion of primary and secondary bile acids per mouse per day in the CTRL and ABX groups at the end of the dietary intervention. Data are presented as mean ± SEM. Asterisks indicate significantly different compared with CTRL. *P < 0.05, **P ≤ 0.001.
DISCUSSION
Emerging evidence points to an association between the gut microbiota and NAFLD (47). However, data on an actual causal role of the gut microbiota in NAFLD are very scarce. Here we found that stimulation of the gut microbiota using GG promoted liver inflammation and fibrosis. Inasmuch as GG is a prebiotic nondigestible carbohydrate and is, thus, expected to act exclusively via its fermentation by the gut bacteria, our results suggest a direct impact of the gut microbiota on the pathogenesis of NAFLD. By contrast, suppression of the gut bacteria using oral antibiotics attenuated liver inflammation and fibrosis. The effects of microbial modulation by GG could be linked to altered circulating and hepatic levels of bile acids, which were shown to induce features of hepatic inflammation and fibrosis. Our data thus suggest that the gut bacteria may be able to influence NAFLD via bile acids.
It should be noted that the stimulation of hepatic inflammation and fibrosis by GG was unrelated to obesity, an important causal factor of NAFLD, as GG suppressed diet-induced weight gain and concomitant glucose intolerance and adipose tissue inflammation. In contrast to GG, RS did not have any impact on NAFLD, probably due to the minor effect of RS on gut microbial composition and levels of fermentation products (48).
Ideally, feeding GG should be combined with the use of antibiotics, thereby allowing direct investigation of the role of the gut microbiota in the effects of GG. However, in our experience it is impossible to combine the use of antibiotics with feeding a diet enriched in dietary fiber. Specifically, we found that giving mice antibiotics while on a high fiber diet quickly led to an intestinal blockage and massive weight loss, forcing us to end the experiment prematurely. This observation underscores the fact that dietary fiber cannot be properly processed in the absence of a well-functioning gut microbiota.
Several mechanisms have been proposed to explain the link between the gut microbiota and host metabolism. These mechanisms mainly revolve around specific microbial metabolites or compounds, including LPS, SCFA, TMA, ethanol, and bile acids, the latter of which are extensively metabolized by the gut microbiota. TMA has been linked to NAFLD by reflecting the microbial conversion and depletion of choline (49). We found significantly higher plasma TMA levels in mice fed GG. Upon absorption, TMA is rapidly oxidized in the liver to TMAO by flavin monooxygenases. Gao et al. (50) showed that dietary TMAO increases adipose tissue mRNA levels and serum levels of monocyte chemoattractant protein-1 (MCP-1) in mice. In addition, TMAO has also been shown to exacerbate atherosclerosis development in mice and is positively linked to cardiovascular disease risk in humans. Importantly, suppression of the gut microbiota using antibiotics lowered plasma TMAO levels and reduced atherosclerosis (9, 45). Furthermore, TMAO was also recently implicated in kidney fibrosis (46). In our study, however, elevated plasma TMAO levels did not seem to be responsible for the aggravation of NALFD in mice fed GG, as feeding mice TMAO for 18 weeks had no effect on any indicators of NALFD or fibrosis, despite markedly elevating circulating and urinary TMAO levels.
In addition to TMA and TMAO, plasma total bile acids were elevated in the GG mice, but not in the RS mice. The parallel reduction in fecal bile acids suggests that bile acids are more efficiently absorbed in the mice fed GG. The increase in plasma bile acid levels was accompanied by elevated hepatic bile acid content and worsening of NAFLD and fibrosis. To investigate whether bile acids could be causally involved in the progression of NAFLD upon GG feeding, mice were fed chow supplemented with taurocholic acid. Taurocholic acid feeding increased plasma levels of a range of different bile acids, which was associated with activation of specific features of NAFLD. These data suggest that elevated plasma and hepatic bile acids are a plausible mechanistic link between changes in gut microbial composition in mice fed GG and worsening of NAFLD. Consistent with this hypothesis, hepatic bile acid levels were found to be elevated in humans with steatohepatitis (51). Furthermore, NAFLD was found to be associated with intestinal dysbiosis and altered fecal bile acid levels in human subjects (52).
One type of bile acid that may be particularly involved in promoting liver injury and NAFLD is deoxycholic acid, levels of which were increased by 4.2-fold in the plasma of mice fed GG. Recently, Yoshimoto et al. (53) linked increased deoxycholic acid levels upon high-fat feeding to the development of NAFLD-related hepatocellular carcinoma. Suppression of the microbiota by antibiotics lowered serum deoxycholic acid levels and mitigated hepatocellular carcinoma development, suggesting that microbial production of deoxycholic acid may have detrimental effects on the liver. The mechanism by which bile acids may promote liver injury and hepatic inflammation and fibrosis may involve enhanced leakage of tight junctions of bile duct epithelial cells causing cholangitis, as well as the activation and proliferation of periductal myofibroblasts leading to periductal fibrosis (54). Furthermore, it is well-established that elevated intrahepatic bile acid levels trigger hepatocyte apoptosis, possibly leading to NASH (55–57). Future studies should address the cellular mechanism(s) by which microbiota-dependent changes in portal and hepatic bile acids may influence NAFLD.
The increase in liver bile acid content in the mice treated with antibiotics may be taken as evidence that bile acids cannot be responsible for the reduced liver injury, inflammation, and fibrosis in these mice. However, it is likely that the increase in liver bile acid content reflects increased bile acid synthesis, as opposed to increased portal delivery of bile acids, which, in fact, was substantially decreased. It is conceivable that the newly synthesized bile acids are immediately secreted and are primarily located in the bile ducts, where they may not inflict liver injury. By contrast, the reduction in portal delivery of bile acids may lead to lower uptake of bile acids by hepatocytes, and thereby attenuate liver injury, inflammation, and fibrosis.
Our results are consistent with previous studies showing that modulation of the gut microbiota influences bile acid levels and metabolism (58–60). These studies have pointed to intestinal FXR signaling and the resultant changes in FGF15 and ceramides as potential intermediates between microbiota-induced changes in bile acids on the one hand, and liver triglycerides and bile acid synthesis on the other hand. Although our data are not inconsistent with a role of intestinal FXR signaling, FGF15, and ceramides, our study highlights the potential role of alterations in portal delivery of bile acids as a causal factor in linking changes in the gut bacteria to altered liver inflammation and fibrosis.
It should be noted that, in contrast to previous data (61–63), we did not find any evidence that the protective effect of antibiotics on the development of NAFLD might be mediated by LPS, as portal LPS levels were not altered upon antibiotic treatment.
In our study, specific gut microbial taxa could be linked to the transition from simple steatosis to NASH and fibrosis. The higher relative abundance of the genera, Bifidobacterium and Prevotella, and of the species, Desulfovibrio C21_c20 and Akkermansia muciniphila, as observed in the colonic luminal content of the GG mice, have not been observed in other mouse studies linking specific gut microbial taxa to the progression of NAFLD. However, those studies used either bile duct ligation (18, 64), a methionine choline-deficient diet (65), or carbon-tetrachloride injections (64) to induce liver injury. Bacteria within the genera, Lactobacillus, Bifidobacterium, and Streptococcus, have been demonstrated to suppress hepatic inflammation in mice and humans (66, 67). In addition to dampening hepatic inflammation, Lactobacillus and Bifidobacterium have also been shown to reduce hepatic fibrosis (68). Furthermore, Bifidobacteria are known to be able to ferment GG (69, 70) and possess active bile salt hydrolases (71). Because unconjugated bile acids are less efficient in the solubilization and absorption of intestinal lipids (71), one possible scenario is that by increasing bile salt hydrolase activity and unconjugated bile acids, the increase in Bifidobacterium may be responsible for elevated fecal lipid excretion.
GG is composed of galactose and mannose residues and is extracted from the seeds of guar beans. It is mainly used as a thickening agent in several food products, including ice cream, sauces, and cheese spreads (72). Although we show that dietary GG promotes the progression of NAFLD, caution should be exercised in extrapolating these data to the human situation because: 1) the dose of GG used substantially exceeds the amounts ingested by humans; and 2) the microbiota composition is vastly different between mice and humans. In our study, we used GG as a model compound to study the effect of modulating the gut microbiota composition on NAFLD.
In conclusion, we found that stimulation of the gut bacteria by GG led to deleterious effects on hepatic inflammation and fibrosis in a mouse model of NAFLD, whereas suppression of the gut bacteria by antibiotics attenuated NAFLD. Overall, we provide evidence of a causal link between disturbances in gut bacteria, bile acids, and NAFLD.
Supplementary Material
Acknowledgments
The authors thank Dr. A. H. Mulder (Rijnstate Hospital, The Netherlands) for excellent advice with histopathological analysis and Martijn Koehorst for the bile acid measurements. The authors are grateful to Dr. J-P. Pais de Barros and H. Choubley (Plateforme de Lipidomique, INSERM UMR1231, LabEx LipSTIC/Endoquant, Dijon, France) for their technical expertise in LPS quantification.
Footnotes
Abbreviations:
- ALT
- alanine aminotransferase
- αSMA
- α smooth muscle actin
- CTRL
- control
- GG
- guar gum
- HFCFD
- high-fat/high-cholesterol/high-fructose diet
- IL
- interleukin
- LPS
- lipopolysaccharide
- NAFLD
- nonalcoholic fatty liver disease
- NASH
- nonalcoholic steatohepatitis
- RS
- resistant starch
- SCFA
- short-chain fatty acid
- TIMP
- tissue inhibitor of metalloproteinases
- TMA
- trimethylamine
- TMAO
- trimethylamine N-oxide
This research was supported by The Netherlands Cardiovascular Research Committee IN-CONTROL Grant (CVON 2012-03) and the Rembrandt Institute for Cardiovascular Research 2012. Trimethylamine and trimethylamine N-oxide analyses were supported by National Institutes of Health Grants HL103866 and HL122283 and Office of Dietary Supplements Grant AA024333. Lipopolysaccharide measurement was supported by Institut National de la Santé et de la Recherche Médicale (INSERM, Centre de Recherches U1231), and the French Government grant managed by the French National Research Agency (ANR-11-LABX-0021) under the program “Investissements d’Avenir”. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Finucane M. M., Stevens G. A., Cowan M. J., Danaei G., Lin J. K., Paciorek C. J., Singh G. M., Gutierrez H. R., Lu Y., Bahalim A. N., et al. . 2011. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet. 377: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ridaura V. K., Faith J. J., Rey F. E., Cheng J., Duncan A. E., Kau A. L., Griffin N. W., Lombard V., Henrissat B., Bain J. R., et al. . 2013. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 341: 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vrieze A., Van Nood E., Holleman F., Salojärvi J., Kootte R. S., Bartelsman J. F. W. M., Dallinga-Thie G. M., Ackermans M. T., Serlie M. J., Oozeer R., et al. . 2012. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 143: 913–916. [DOI] [PubMed] [Google Scholar]
- 4.Lam Y. Y., Ha C. W. Y., Campbell C. R., Mitchell A. J., Dinudom A., Oscarsson J., Cook D. I., Hunt N. H., Caterson I. D., Holmes A. J., et al. . 2012. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One. 7: e34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janssen A. W. F., and Kersten S.. 2015. The role of the gut microbiota in metabolic health. FASEB J. 29: 3111–3123. [DOI] [PubMed] [Google Scholar]
- 6.Sommer F., and Bäckhed F.. 2013. The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol. 11: 227–238. [DOI] [PubMed] [Google Scholar]
- 7.Clemente J. C., Ursell L. K., Parfrey L. W., and Knight R.. 2012. The impact of the gut microbiota on human health: an integrative view. Cell. 148: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biedermann L., and Rogler G.. 2015. The intestinal microbiota: its role in health and disease. Eur. J. Pediatr. 174: 151–167. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z., Klipfell E., Bennett B. J., Koeth R., Levison B. S., Dugar B., Feldstein A. E., Britt E. B., Fu X., Chung Y.-M., et al. . 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 472: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janssen A. W. F., and Kersten S.. 2017. Potential mediators linking gut bacteria to metabolic health: a critical view. J. Physiol. 595: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuppan D., and Schattenberg J. M.. 2013. Non-alcoholic steatohepatitis: pathogenesis and novel therapeutic approaches. J. Gastroenterol. Hepatol. 28(Suppl. 1): 68–76. [DOI] [PubMed] [Google Scholar]
- 12.Tilg H., and Moschen A. R.. 2010. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology. 52: 1836–1846. [DOI] [PubMed] [Google Scholar]
- 13.Nassir F., and Ibdah J.. 2014. Role of mitochondria in nonalcoholic fatty liver disease. Int. J. Mol. Sci. 15: 8713–8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang W., Wu N., Wang X., Chi Y., Zhang Y., Qiu X., Hu Y., Li J., and Liu Y.. 2015. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci. Rep. 5: 8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouzaki M., Comelli E. M., Arendt B. M., Bonengel J., Fung S. K., Fischer S. E., Mcgilvray I. D., and Allard J. P.. 2013. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 58: 120–127. [DOI] [PubMed] [Google Scholar]
- 16.Jiang C., Xie C., Li F., Zhang L., Nichols R. G., Krausz K. W., Cai J., Qi Y., Fang Z., Takahashi S., et al. . 2015. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J. Clin. Invest. 125: 386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henao-Mejia J., Elinav E., Jin C., Hao L., Mehal W. Z., Strowig T., Thaiss C. A., Kau A. L., Eisenbarth S. C., Jurczak M. J., et al. . 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 482: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Minicis S., Rychlicki C., Agostinelli L., Saccomanno S., Candelaresi C., Trozzi L., Mingarelli E., Facinelli B., Magi G., Palmieri C., et al. . 2014. Dysbiosis contributes to fibrogenesis in the course of chronic liver injury in mice. Hepatology. 59: 1738–1749. [DOI] [PubMed] [Google Scholar]
- 19.Nomura K., and Yamanouchi T.. 2012. The role of fructose-enriched diets in mechanisms of nonalcoholic fatty liver disease. J. Nutr. Biochem. 23: 203–208. [DOI] [PubMed] [Google Scholar]
- 20.Song P., Zhang Y., and Klaassen C. D.. 2011. Dose-response of five bile acids on serum and liver bile acid concentrations and hepatotoxicty in mice. Toxicol. Sci. 123: 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., and Medzhitov R.. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 118: 229–241. [DOI] [PubMed] [Google Scholar]
- 22.Bieghs V., Wouters K., van Gorp P. J., Gijbels M. J. J., de Winther M. P. J., Binder C. J., Lütjohann D., Febbraio M., Moore K. J., van Bilsen M., et al. . 2010. Role of scavenger receptor A and CD36 in diet-induced nonalcoholic steatohepatitis in hyperlipidemic mice. Gastroenterology. 138: 2477–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy R. A., van Montfoort A. P. A., Dikkers A., van Echten-Arends J., Homminga I., Land J. A., Hoek A., and Tietge U. J. F.. 2015. Presence of bile acids in human follicular fluid and their relation with embryo development in modified natural cycle IVF. Hum. Reprod. 30: 1102–1109. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z., Levison B. S., Hazen J. E., Donahue L., Li X. M., and Hazen S. L.. 2014. Measurement of trimethylamine-N-oxide by stable isotope dilution liquid chromatography tandem mass spectrometry. Anal. Biochem. 455: 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koeth R. A., Levison B. S., Culley M. K., Buffa J. A., Wang Z., Gregory J. C., Org E., Wu Y., Li L., Smith J. D., et al. . 2014. γ-Butyrobetaine Is a proatherogenic intermediate in gut microbial metabolism of L–carnitine to TMAO. Cell Metab. 20: 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Govers M. J., and Van der Meet R.. 1993. Effects of dietary calcium and phosphate on the intestinal interactions between calcium, phosphate, fatty acids, and bile acids. Gut. 34: 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Meer H., Boehm G., Stellaard F., Vriesema A., Knol J., Havinga R., Sauer P. J., and Verkade H. J.. 2008. Prebiotic oligosaccharides and the enterohepatic circulation of bile salts in rats. Am. J. Physiol. Gastrointest. Liver Physiol. 294: G540–G547. [DOI] [PubMed] [Google Scholar]
- 28.Bolstad B. M., Irizarry R. A., Astrand M., and Speed T. P.. 2003. A comparison of normalization metholds for high density oligonucleotide array data based on variance and bias. Bioinformatics. 19: 185–193. [DOI] [PubMed] [Google Scholar]
- 29.Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., and Speed T. P.. 2003. Summaries of affymetrix genechip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dai M., Wang P., Boyd A. D., Kostov G., Athey B., Jones E. G., Bunney W. E., Myers R. M., Speed T. P., Akil H., et al. . 2005. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sartor M. A., Tomlinson C. R., Wesselkamper S. C., Sivaganesan S., Leikauf G. D., and Medvedovic M.. 2006. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 7: 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storey J. D., and Tibshirani R.. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jamall I. S., Finelli V. N., and Que Hee S. S.. 1981. A simple method to determine nanogram levels of 4-hydroxyproline in biological tissues. Anal. Biochem. 112: 70–75. [DOI] [PubMed] [Google Scholar]
- 34.Gevers D., Kugathasan S., Denson L. A., Vázquez-Baeza Y., Van Treuren W., Ren B., Schwager E., Knights D., Song S. J., Yassour M., et al. . 2014. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 15: 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., et al. . 2010. QIIME allows analysis of high- throughput community sequencing data. Nat. Methods. 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., and Huttenhower C.. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki M. T., Taylor L. T., and DeLong E. F.. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66: 4605–4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisburg W. G., Barns S. M., Pelletier D. A., and Lane D. J.. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pryce T. M., Palladino S., Kay I. D., and Coombs G. W.. 2003. Rapid identification of fungi by sequencing the ITSl and ITS2 regions using an automated capillary electrophoresis system. Med. Mycol. 41: 369–381. [DOI] [PubMed] [Google Scholar]
- 40.Haenen D., Souza C., Zhang J., Koopmans S. J., Bosch G., Vervoort J., Gerrits W. J. J., Kemp B., Smidt H., and Mu M.. 2013. Resistant starch induces catabolic but suppresses immune and cell division pathways and changes the microbiome in the proximal colon of male pigs. J. Nutr. 143: 1889–1898. [DOI] [PubMed] [Google Scholar]
- 41.Charlton M., Krishnan A., Viker K., Sanderson S., Cazanave S., McConico A., Masuoko H., and Gores G.. 2011. Fast food diet mouse: novel small animal model of NASH with ballooning, progressive fibrosis, and high physiological fidelity to the human condition. Am. J. Physiol. Gastrointest. Liver Physiol. 301: G825–G834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawanishi N., Yano H., Mizokami T., Takahashi M., Oyanagi E., and Suzuki K.. 2012. Exercise training attenuates hepatic inflammation, fibrosis and macrophage infiltration during diet induced-obesity in mice. Brain Behav. Immun. 26: 931–941. [DOI] [PubMed] [Google Scholar]
- 43.Savard C., Tartaglione E. V., Kuver R., Haigh W. G., Farrell G. C., Subramanian S., Chait A., Yeh M. M., Quinn L. S., and Ioannou G. N.. 2013. Synergistic interaction of dietary cholesterol and dietary fat in inducing experimental steatohepatitis. Hepatology. 57: 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bashiardes S., Shapiro H., Rozin S., Shibolet O., and Elinav E.. 2016. Non-alcoholic fatty liver and the gut microbiota. Mol. Metab. 5: 782–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koeth R. A., Wang Z., Levison B. S., Buffa J. A., Org E., Sheehy B. T., Britt E. B., Fu X., Wu Y., Li L., et al. . 2013. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19: 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang W. H., Wang Z., Kennedy D. J., Wu Y., Buffa J. A., Agatisa-Boyle B., Li X. S., Levison B. S., and Hazen S. L.. 2015. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 116: 448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau E., Carvalho D., and Freitas P.. 2015. Gut microbiota: association with NAFLD and metabolic disturbances. BioMed Res. Int. 2015: 979515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lange K., Hugenholtz F., Jonathan M. C., Schols H. A., Kleerebezem M., Smidt H., Müller M., and Hooiveld G. J.. 2015. Comparison of the effects of five dietary fibers on mucosal transcriptional profiles, and luminal microbiota composition and SCFA concentrations in murine colon. Mol. Nutr. Food Res. 59: 1590–1602. [DOI] [PubMed] [Google Scholar]
- 49.Dumas M-E., Barton R. H., Toye A., Cloarec O., Blancher C., Rothwell A., Fearnside J., Tatoud R., Blanc V., Lindon J. C., et al. . 2006. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. USA. 103: 12511–12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao X., Liu X., Xu J., Xue C., Xue Y., and Wang Y.. 2014. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J. Biosci. Bioeng. 118: 476–481. [DOI] [PubMed] [Google Scholar]
- 51.Aranha M. M., Cortez-Pinto H., Costa A., da Silva I. B. M., Camilo M. E., de Moura M. C., and Rodrigues C. M. P.. 2008. Bile acid levels are increased in the liver of patients with steatohepatitis. Eur. J. Gastroenterol. Hepatol. 20: 519–525. [DOI] [PubMed] [Google Scholar]
- 52.Mouzaki M., Wang A. Y., Bandsma R., Comelli E. M., Arendt B. M., Zhang L., Fung S., Fischer S. E., McGilvray I. G., and Allard J. P.. 2016. Bile acids and dysbiosis in non-alcoholic fatty liver disease. PLoS One. 11: e0151829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshimoto S., Loo T. M., Atarashi K., Kanda H., Sato S., Oyadomari S., Iwakura Y., Oshima K., Morita H., Hattori M., et al. . 2013. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 499: 97–101. [Erratum. 2014. Nature. 506: 396.] [DOI] [PubMed] [Google Scholar]
- 54.Fickert P., Fuchsbichler A., Marschall H-U., Wagner M., Zollner G., Krause R., Zatloukal K., Jaeschke H., Denk H., and Trauner M.. 2006. Lithocholic acid feeding induces segmental bile duct obstruction and destructive cholangitis in mice. Am. J. Pathol. 168: 410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woudenberg-Vrenken T. E., Conde de la Rosa L., Buist-Homan M., Faber K. N., and Moshage H.. 2013. Metformin protects rat hepatocytes against bile acid-induced apoptosis. PLoS One. 8: e71773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sodeman T., Bronk S. F., Roberts P. J., Miyoshi H., and Gores G. J.. 2000. Bile salts mediate hepatocyte apoptosis by increasing cell surface trafficking of Fas. Am. J. Physiol. Gastrointest. Liver Physiol. 278: G992–G999. [DOI] [PubMed] [Google Scholar]
- 57.Higuchi H., Bronk S. F., Takikawa Y., Werneburg N., Takimoto R., El-deiry W., and Gores G. J.. 2001. The bile acid glycochenodeoxycholate induces TRAIL-receptor 2 / DR5 expression and apoptosis. J. Biol. Chem. 276: 38610–38618. [DOI] [PubMed] [Google Scholar]
- 58.Swann J. R., Want E. J., Geier F. M., Spagou K., Wilson I. D., Sidaway J. E., Nicholson J. K., and Holmes E.. 2011. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl. Acad. Sci. USA. 108: 4523–4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sayin S. I., Wahlström A., Felin J., Jäntti S., Marschall H-U., Bamberg K., Angelin B., Hyötyläinen T., Orešič M., and Bäckhed F.. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17: 225–235. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y., Limaye P. B., Renaud H. J., and Klaassen C. D.. 2014. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol. Appl. Pharmacol. 277: 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rahman K., Desai C., Iyer S. S., Thorn N. E., Kumar P., Liu Y., Smith T., Neish A. S., Li H., Tan S., et al. . 2016. Loss of junctional adhesion molecule A promotes severe steatohepatitis in mice on a diet high in saturated fat, fructose, and cholesterol. Gastroenterology. 151: 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carvalho B. M., Guadagnini D., Tsukumo D. M. L., Schenka A. A., Latuf-Filho P., Vassallo J., Dias J. C., Kubota L. T., Carvalheira J. B. C., and Saad M. J. A.. 2012. Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia. 55: 2823–2834. [DOI] [PubMed] [Google Scholar]
- 63.Douhara A., Moriya K., Yoshiji H., Noguchi R., Namisaki T., Kitade M., Kaji K., Aihara Y., Nishimura N., Takeda K., et al. . 2015. Reduction of endotoxin attenuates liver fibrosis through suppression of hepatic stellate cell activation and remission of intestinal permeability in a rat non-alcoholic steatohepatitis model. Mol. Med. Rep. 11: 1693–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fouts D. E., Torralba M., Nelson K. E., Brenner D. A., and Schnabl B.. 2012. Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. J. Hepatol. 56: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henao-Mejia J., Elinav E., Thaiss C. A., and Flavell R. A.. 2013. The intestinal microbiota in chronic liver disease. Adv. Immunol. 117: 73–97. [DOI] [PubMed] [Google Scholar]
- 66.Li Z., Yang S., Lin H., Huang J., Watkins P. A., Moser A. B., DeSimone C., Song X. Y., and Diehl A. M.. 2003. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology. 37: 343–350. [DOI] [PubMed] [Google Scholar]
- 67.Eslamparast T., Poustchi H., Zamani F., Sharafkhah M., Malekzadeh R., and Hekmatdoost A.. 2014. Synbiotic supplementation in nonalcoholic fatty liver disease : a randomized, double-blind, placebo-controlled pilot study. Am. J. Clin. Nutr. 99: 535–542. [DOI] [PubMed] [Google Scholar]
- 68.Okubo H., Sakoda H., Kushiyama A., Fujishiro M., Nakatsu Y., Fukushima T., Matsunaga Y., Kamata H., Asahara T., Yoshida Y., et al. . 2013. Lactobacillus casei strain Shirota protects against nonalcoholic steatohepatitis development in a rodent model. Am. J. Physiol. Gastrointest. Liver Physiol. 305: G911–G918. [DOI] [PubMed] [Google Scholar]
- 69.Noack J., Kleessen B., Proll J., Dongowski G., and Blaut M.. 1998. Dietary guar gum and pectin stimulate intestinal microbial polyamine synthesis in rats. J. Nutr. 128: 1385–1391. [DOI] [PubMed] [Google Scholar]
- 70.Ohashi Y., Sumitani K., Tokunaga M., Ishihara N., Okubo T., and Fujisawa T.. 2015. Consumption of partially hydrolysed guar gum stimulates Bifidobacteria and butyrate-producing bacteria in the human large intestine. Benef. Microbes. 6: 451–455. [DOI] [PubMed] [Google Scholar]
- 71.Begley M., Hill C., and Gahan C. G. M.. 2006. Bile salt hydrolase activity in probiotics bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72: 1729–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mudgil D., Barak S., and Khatkar B. S.. 2014. Guar gum: processing, properties and food applications—a review. J. Food Sci. Technol. 51: 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.