Abstract
Purpose
To evaluate DRAMMS, an attribute-based deformable registration algorithm, compared to other intensity-based algorithms, for longitudinal breast MRI registration, and to show its applicability in quantifying tumor changes over the course of neoadjuvant chemotherapy.
Methods
Breast magnetic resonance images from 14 women undergoing neoadjuvant chemotherapy were analyzed. The accuracy of DRAMMS versus five intensity-based deformable registration methods was evaluated based on 2,380 landmarks independently annotated by two experts, for the entire image volume, different image subregions, and patient subgroups. The registration method with the smallest landmark error was used to quantify tumor changes, by calculating the Jacobian determinant maps of the registration deformation.
Results
DRAMMS had the smallest landmark errors (6.05 ± 4.86 mm), followed by the intensity-based methods CC-FFD (8.07 ± 3.86 mm), NMI-FFD (8.21 ± 3.81 mm), SSD-FFD (9.46 ± 4.55 mm), Demons (10.76 ± 6.01 mm), and Diffeomorphic Demons (10.82 ± 6.11 mm). Results show that registration accuracy also depends on tumor versus normal tissue regions and different patient subgroups.
Conclusions
The DRAMMS deformable registration method, driven by attribute-matching and mutual-saliency, can register longitudinal breast magnetic resonance images with a higher accuracy than several intensity-matching methods included in this article. As such, it could be valuable for more accurately quantifying heterogeneous tumor changes as a marker of response to treatment.
Keywords: deformable image registration, breast cancer, longitudinal breast MRI, tumor changes, evaluation, treatment
INTRODUCTION
Breast cancer is the second most frequent cancer and the second leading cause of death for women in the United States (1). A critical issue in the treatment of locally advanced breast cancer is how to effectively monitor tumor changes during the course of neoadjuvant chemotherapy, and predict the long-term pathologic response. This is needed so that clinicians can make necessary treatment adjustments and optimize treatment effects (2,3). To do so, clinicians often refer to longitudinal breast magnetic resonance (MR) images, acquired in multiple visits, days or weeks apart (a series of longitudinal breast MR images can be found for a representative breast cancer patient in the first row of Fig. 7). Here, a central problem is how to accurately quantify tumor changes over time, and more importantly, how to effectively reveal heterogeneous changes in various subregions within tumors, rather than tumors as a whole, since the heterogeneity may be more indicative of long-term pathologic response (11–16). Toward this goal, recent research has increasingly focused on image registration techniques (17,18). In this article, we evaluate the performance of six image registration methods specifically for the registration of longitudinal breast MR images. One primary objective is to evaluate an attribute-based deformable registration method, DRAMMS (19), and compare it with other intensity-based registration methods. We also demonstrate the feasibility of applying registration as a means to quantify tumor changes over time, which can be used as a potential marker for a breast cancer patient’s response to neoadjuvant chemotherapy.
FIG. 7.
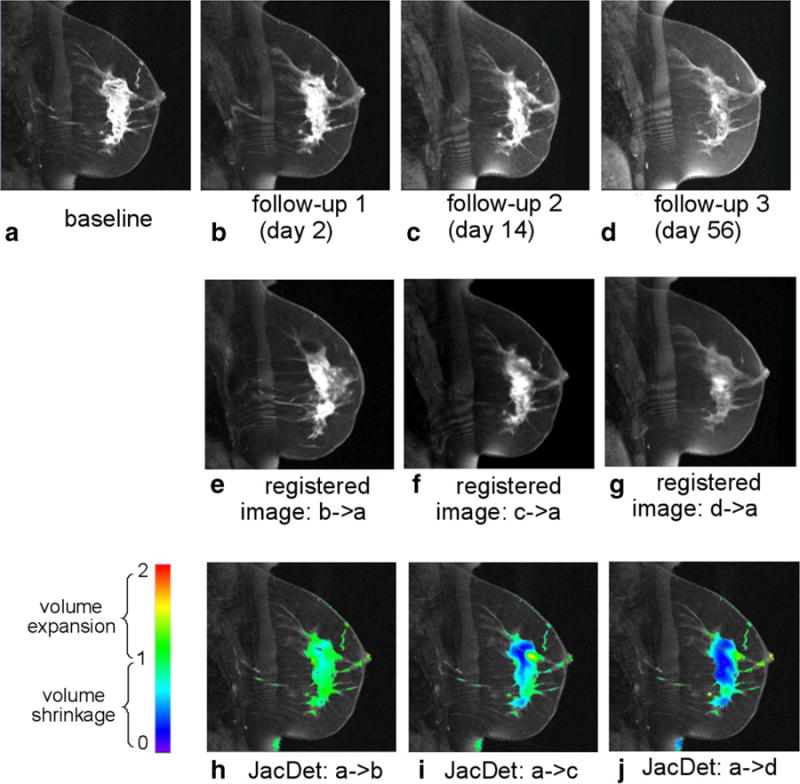
Quantification of the voxelwise longitudinal breast cancer change by the DRAMMS registration algorithm. Results shown in this figure are from an example pathologic partial responder (pPR) patient. Jacobian determinants in the tumor regions are overlaid on the baseline image in the bottom row.
Longitudinal Breast MR Image Registration: Definition and Necessity
By definition, image registration is the process of finding an optimal transformation that spatially aligns two or more images. In the context of longitudinal images, deformable registration can capture how anatomical structures deform over time. This concept is shown in Figure 1. In this figure, a follow-up image (Fig. 1b) is registered to the baseline image (Fig. 1a). The resultant deformation is visualized in Figure 1(d). The deformation is further quantitatively analyzed to derive the Jacobian determinant map, as shown in Figure 1e. The Jacobian determinant value at a voxel indicates the volumetric change ratio (follow-up/baseline): >1 for expansion, <1 for shrinkage, and = 1 for volume preservation. Therefore, if the solid white plate in Figure 1 symbolizes a changing tumor, then the Jacobian determinant map in Figure 1e shows that the tumor has shrunk from the baseline to the follow-up images, while at the same time the neighborhood has expanded to fill into the region that was once tumorous. This conceptually demonstrates that the deformable image registration can quantify longitudinal changes, and that the quantification of changes agrees with the human interpretation.
FIG. 1.
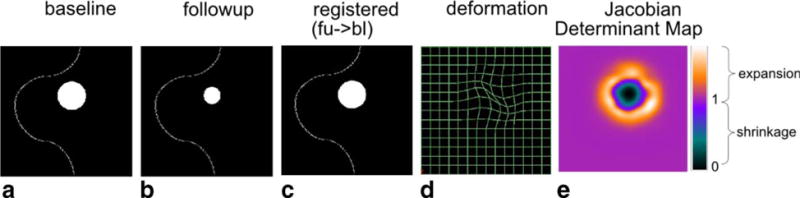
A conceptual illustration of using deformable registration to quantify the volumetric changes of tumors and the surrounding tissue in longitudinal images. The solid white plate symbolizes a tumor that has shrunk at the follow-up visit.
Deformable registration can also reveal the spatial heterogeneity of changes at the voxel level (17,18). In contrast, the standard RECIST rule (20,21) or other segmentation-based approaches (22,23) could only tell how the tumors change as a whole. Indeed, tumors often change heterogeneously in space (24–29), meaning that different regions within a tumor may respond differently to the treatment, and hence they may change heterogeneously over time. Several studies have shown that such heterogeneity within tumors (30–33) and the surrounding tissue (34) can help predict the long-term pathologic response to treatment.
Need for Evaluating Registration Performance for Longitudinal Breast MR Images
Despite its importance, longitudinal registration of breast MRI has been less studied, and the registration performance is less understood than that for other organs [e.g., brain (35,36), lung (37), heart (38), and liver (39,40)].
Prior studies for breast image registration were either on two-dimensional (2-D) mammographic images (41–47), or on “short-interval” longitudinal breast MR images, which are acquired seconds or minutes apart during one visit [e.g., (48–54)]. There, registration is used mainly for motion correction (55–62). In contrast, to quantify the heterogeneous tumor changes, we are dealing with “long-interval” longitudinal breast MR images, which are acquired days or weeks apart in multiple visits. Few registration methods exist for this task. Moreover, a comprehensive evaluation of those methods remains an unexplored topic, which motivates our work in this article.
Outline and Contributions
This article evaluates six deformable registration methods that have been previously reported for the registration of long-interval longitudinal breast MR images. Hereafter, we will use the term “longitudinal breast MR images” as short for “long-interval longitudinal breast MR images,” unless otherwise specified. Out of those six methods, DRAMMS (19) is an attribute-based method (i.e., it finds voxel correspondences by the high-dimensional attributes extracted from the neighborhoods of voxels); while, in contrast, the other five are intensity-based methods (i.e., they find voxel correspondences by maximizing some image similarities defined on the intensity or intensity distributions). A primary purpose of our study is to evaluate whether the use of attributes in DRAMMS for characterizing voxels can help improve the registration accuracy over the intensity-based methods. A second purpose is to show preliminary results on using registration to quantify tumor changes over time as a measure of patient response to treatment. As such, our work presents the following contributions:
We evaluate six deformable registration methods specifically for longitudinal breast MR images. They have all been reported as being used for this specific task of longitudinal breast MRI registration. Within this context, we examine the advantages of different registration approaches, with an emphasis on the comparison between attribute- and intensity-based registration algorithms. We also point out potential areas to improve in the future.
We quantitatively measure the performance of registration methods based on two breast radiologists independently annotating a total of 2,380 landmarks in longitudinal MRI scans of 14 breast cancer patients.
We further separately examine registration errors in tumor versus normal tissue regions. Studies have shown that both types of regions can contribute to the prediction of long-term response to treatment (34). However, little is known about whether we should use them differently. This largely depends on whether the registration performance, and hence the subsequently registration-quantified changes, are different between tumor versus normal tissue regions, which has not yet been investigated in the literature.
Finally, we quantitatively examine whether the registration performance is different for patient subgroups having different pathologic responses to treatment. The hypothesis is that, tumors may change more in patients having pathologic complete responses than patients having pathologic partial responses, thereby posing a higher level of challenge to registration.
The rest of this article is organized as follows. In Methods section, we describe our evaluation protocol. In Results section, we show the evaluation results, and show preliminary results of using deformable registration to quantify tumor changes. Finally, Conclusion section discusses and concludes this article.
METHODS
Protocol to Evaluate Registration Accuracy
This section presents the evaluation protocol. In the next four subsections, we will describe our dataset, the registration methods to be evaluated, the parameter settings for those registration methods, and the criterion we used to evaluate the registration accuracy.
Dataset
We have retrospectively collected longitudinal breast MR images (1.5T Siemens Sonata scanner) for 14 women with biopsy-proven T2–3 breast tumors from the ACRIN 6657 I-SPY trial (63,64). All women in this trial underwent the standard neoadjuvant chemotherapy, which, at the time of the study, consisted of four cycles of adriamycin/cytoxan, followed by four cycles of taxotere. At the end of the 3–4 months of chemotherapy, they were evaluated by a pathological analysis. Those who showed the absence of any residual invasive cancer in the breast and the absence of any metastatic cells in the regional lymph nodes were defined as pathologic complete responders (pCR; (65,66)), otherwise as pathologic partial responders (pPR). In our dataset, eight women were classified as pCR and six as pPR.
Imaging parameters were: FOV 18–20 cm, image size 256 × 256 × 64, voxel size 0.70 × 0.70 × 2.0 mm3, pulse repetition time (TR) = 27.0 ms, echo time (TE) = 4.76 ms, flip angle = 45°. Pre-gadodiamide imaging was performed followed by immediate post-gadolinium images (at 2 min) and delayed post-gadodiamide images (at 7 min). The baseline image was the image obtained right before the first chemotherapy. We registered the follow-up image after two rounds of chemotherapy to the baseline image for each patient (average 31 ± 21 days apart). Table 1 lists the information of each patient in our dataset.
Table 1.
Lists of Cases in the Pathologic Complete Responder (pCR) and Partial Responder (pPR) Subgroups in Our Study
| #days (FU-BL) | #LM (tumor) | #LM (normal) | #LM (all) | |
|---|---|---|---|---|
| pPR group | ||||
| Patient 1 | 14 | 63 | 59 | 122 |
| Patient 2 | 29 | 25 | 55 | 80 |
| Patient 3 | 13 | 64 | 60 | 124 |
| Patient 4 | 14 | 22 | 45 | 67 |
| Patient 5 | 14 | 90 | 16 | 106 |
| Patient 6 | 56 | 53 | 55 | 108 |
| Mean ± stdev | 23 ± 17 | 53 ± 26 | 48 ± 17 | 101 ± 23 |
| pCR group | ||||
| Patient 7 | 61 | 42 | 30 | 72 |
| Patient 8 | 14 | 14 | 48 | 62 |
| Patient 9 | 60 | 16 | 45 | 61 |
| Patient 10 | 56 | 8 | 19 | 27 |
| Patient 11 | 14 | 22 | 81 | 103 |
| Patient 12 | 55 | 41 | 20 | 61 |
| Patient 13 | 21 | 51 | 25 | 76 |
| Patient 14 | 14 | 31 | 87 | 118 |
| Mean ± stdev | 37 ± 23 | 28 ± 15 | 44 ± 27 | 73 ± 28 |
| All patients | ||||
| Mean ± stdev | 31 ± 21 | 39 ± 23 | 46 ± 22 | 85 ± 29 |
The time interval between the follow-up and baseline images, and the number of manually annotated landmarks in tumor and normal tissue regions are listed for each patient. (FU: follow-up; BL: baseline; LM: landmarks.)
Two breast imaging radiologists (SW and EC) annotated landmarks independently in the images, blinded to the patient response outcome. First, they agreed on a common set of anatomical/geometric landmarks in the baseline images. Then, they independently defined in the follow-up images the landmarks, which, according to their expert knowledge, were corresponding to the same set of landmarks in the baseline images. The baseline and follow-up images were displayed side-by-side to facilitate expert annotations. The “New VOI”—“Draw Point VOI” function in the MIPAV software (67) was used by the experts to annotate the landmark correspondences (VOI stands for volume of interest). When an expert chose a landmark location by a click of the mouse, the “Draw Point VOI” function placed a cross in the image at the same location. At the same time, a number was associated with the cross, indexing the order in which the landmarks were annotated (i.e., Fig. 2). Locations of the same index numbers were considered corresponding landmark locations defined by experts in the two images. In general, landmarks were scattered in the image space to measure the accuracy of registration in various regions. Landmarks were usually located at nipples, breast boundaries, chest walls, internal milk ducts, vessels, glandular structures as well as tumors. As such, a total of 2,380 landmark pairs (85 ± 29 per patient) were annotated and labeled as being within tumor or normal tissue regions. As Table 1 and Figure 2 show, patients in the pCR and pPR subgroups have similar numbers of landmarks in the normal tissue regions, but pCR patients have significantly fewer landmarks in the tumor regions, highlighting the larger changes in tumors and hence an increased level of ambiguity in finding corresponding landmarks.
FIG. 2.
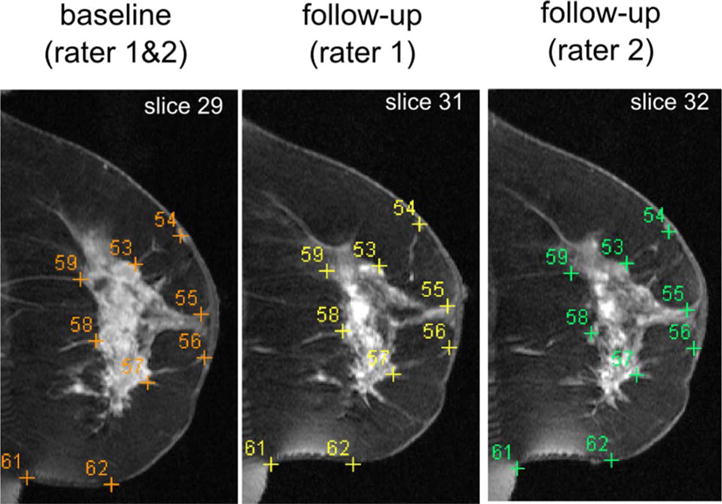
Example landmarks annotated by human experts in a representative patient (patient #1). Landmarks with the same indexing numbers in different images correspond to each other, according to the expert annotations. Note that the images have been manually cropped for the display purpose. In the automated registration, however, we used the entire images as shown in the first rows in Figs. 7 and 8, where no manual or automated cropping was involved.
Figure 2 shows some example expert-annotated landmark correspondences in a representative subject (i.e., patient #1). Landmarks of the same indexing number in the different images correspond to each other according to expert annotations. Please note that, for the same set of landmarks in the baseline image (in the left panel of the figure), the annotated corresponding landmark locations by expert 1 (in the middle panel) and expert 2 (in the right panel) were slightly different. Such differences can occur in the in-plane x–y locations, and also in the z-direction (see, e.g., the different slice numbers at the upper right corners of subfigures). The distribution of interexpert landmark differences can be found in the box-plots shown in Figure 4. In general, experts differ by 0–5 mm at a majority of (>80%) landmark locations, and they differ by more than 10 mm only at few (<5%) landmark locations.
FIG. 4.
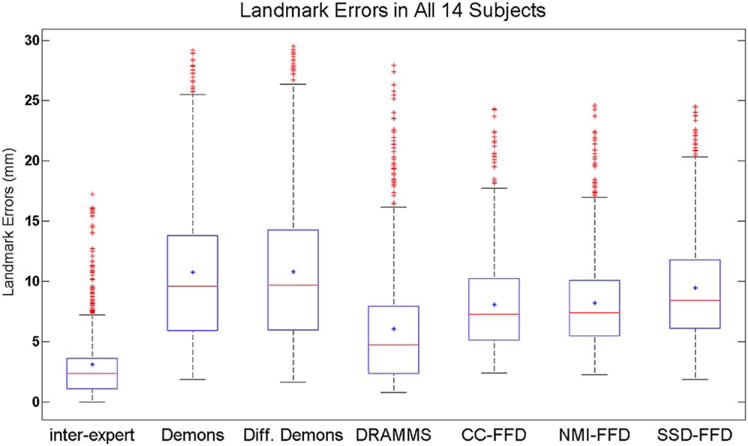
The overall landmark errors in all 14 subjects, for interexpert errors and algorithm-to-expert errors.
All images were converted into the NIfTI image format. Before registration, they all went through the N3-based bias field correction (using the default parameters; 68). Histograms of the follow-up images were matched to histograms of the baseline images, using the histogram matching module of the MIPAV software (67). This was to reduce intensity differences caused by imaging artifacts or by the imaging sequences used at different times.
Registration Methods to be Evaluated
The registration methods included in this evaluation work satisfied two criteria:
They were previously reported in the literature to register (long-interval) longitudinal breast MR images for measuring the volumetric changes of tumors, and their corresponding implementation parameters were disclosed.
Their software packages are publicly available, therefore, allowing a wide use of them such as in this study.
Through our search of the literature, we found six registration algorithms satisfying these criteria. They have been implemented in three software packages: Demons (69,70), free form deformation (FFD; (71)), and DRAMMS (19). Demons was directly used in (17) and slightly modified in (72) to register breast MRIs acquired weeks or months apart (73). An FFD variant version (74) was used in (18) to register longitudinal breast MR images with known synthetic tumor changes. DRAMMS was used in (19,75) but was compared to FFD for longitudinal breast MRI registration only qualitatively. Below we summarize the registration methods evaluated in our study.
(Additive) Demons and Diffeomorphic Demons: Demons (69,70) is an intensity-based deformable registration framework. It considers deformable image registration as a nonparametric diffusion process. It matches voxels to their correspondences according to local intensity characterizations. Using the intensity difference as the similarity metric, a force is computed from the optical flow equations to push voxels by some velocity that is iteratively combined with the total displacement (initially zero), which is later smoothed by Gaussian filters for regularization. (Additive) Demons (69) adds the velocity field to the current deformation in each iteration, whereas Diffeomorphic Demons (70) composes the deformation with the exponential of the velocity field, enforcing diffeomorphism. In this article, both (Additive) Demons and Diffeomorphic Demons were included in the evaluation. They are both available in the Demons software, which can be downloaded from http://www.insight-journal.org/browse/publication/154.
FFD and its variants: FFD is a geometric transformation model introduced to deformable image registration in 1999 (71) and widely used thereafter. It is often used in combination with intensity-based matching criteria, hence intensity-based registration algorithms. In the FFD model, a regular grid of “control points” is superimposed on top of the dense image lattice. The main idea is that the movement of an image voxel is a smooth, cubic-B-spline-based interpolation of the displacements of the neighboring control points. Therefore, the task of finding movements at each voxel is translated into finding the displacements at a much smaller number of regularly spaced control points. In contrast to landmark-based methods, FFD has three properties: (1) control points are regularly spaced in the image, providing guidance throughout the image domain; (2) deformation is regularized by the cubic-B-spline-based interpolation; and (3) a landmark moves by finding its own correspondence, whereas a control point moves by finding the most likely corresponding patches for the image patches it represents. The FFD transformation model can be combined with similarity metrics such as the normalized mutual information (NMI), the correlation coefficient (CC), and the sum of squared difference (SSD) of the two images to be registered. NMI-FFD, CC-FFD, and SSD-FFD were all included in our evaluation. The software package, known as IRTK, is available at http://www.doc.ic.ac.uk/dr/software/.
DRAMMS: DRAMMS (19) is an attribute-based deformable image registration algorithm. It characterizes voxels by the high-dimensional Gabor texture attributes extracted from the multiscale and multiorientation neighborhoods of voxels. This has the potential to increase the accuracy and reliability of voxelwise matching, especially in the presence of intensity inhomogeneity and contrast differences between longitudinal scans. Besides, DRAMMS uses a newly developed mutual-saliency function, which automatically quantifies the matching reliability at the voxel level. Based on this function, DRAMMS guides the registration with those voxels/regions having higher confidence to establish correspondences across images. DRAMMS uses the cubic-B-spline FFD transformation model ((71); described above) and the discrete optimization strategy (76,77). The DRAMMS software package is available at http://www.cbica.upenn.edu/sbia/software/dramms/.
Parameter Settings in Registration Methods
For (Additive) Demons and Diffeomorphic Demons, the key parameters are (i) the Gaussian smoothing kernel (σ1) of the velocity field; and (ii) the Gaussian smoothing kernel (σ2) after having combined the velocity field into the overall deformation field. Larger smoothing kernels σ1; σ2 lead to smoother deformations. Study (17) used Demons directly to recover breast changes in longitudinal MR images 3 months apart, which is fairly similar to our dataset with the same objective. In their study, Demons was used with σ1 = 8 and σ2 = 1. In another study (73), which registered breast MR images weeks apart, an algorithm similar to Demons was used with parameter σ1 = 7. In a related study (78) registering breast elastography, which has image sizes and voxel sizes similar to breast MRI, the authors used σ2 = 1.5. Since the parameters in all three independent studies were similar, we also followed their settings and chose the Demons parameters at σ1 = 8 and σ2 = 1.5. In addition, we allowed the maximum number of iterations in three image resolutions to be 30 × 20 × 10, which doubles the default maximum number of iterations and was used in a recent studies (35,80) to ensure the convergence of the program.
For FFD and its variants, the key parameters are (i) the control point spacing δ, with a larger spacing capturing more global deformations; and (ii) the regularization weight λ, with larger weights providing smoother deformations. We set δ = 10 mm and λ = 0.01 following the original FFD paper (71), which, together with other studies from the same group [e.g., (48,60)], found this set of parameters to perform stably and well suit the breast MR image registration task.
For DRAMMS, most parameters are automatically adaptive to the input images (i.e., size, contrast, and histogram), and hence do not need extensive tuning. One parameter that may need to be tuned is the regularization weight g, which is usually set between 0 and 1, with higher weights for smoother deformations, and even greater than 1 for very smooth deformations. We set g = 0.3, which led to slightly smoother deformations than using the default value g = 0.2. This was also the parameter used in its original works (19,75) for longitudinal breast MR image registration experiments.
Note that the parameters mentioned above were not set arbitrarily. Usually, the authors of the aforementioned studies tried a wide range of parameter settings, and reported the ones that led to the most stable and/or most accurate alignment of breast MR images.
Evaluation Criterion for Registration Accuracy
Evaluating the accuracy of registration is a difficult task in general. This is mainly due to the lack of the ground-truth deformation at each and every voxel. In other registration evaluation studies (e.g., for brain, lung, or short-interval longitudinal breast MR images), existing work has used simulated images/deformations [e.g., (41,44,45,51,79)], expert-annotated regions of interest [ROI; e.g., (35,38,43)], or expert-annotated landmarks [e.g., (43,73)]. While simulations provide ground-truth deformations, they do not offer fully realistic deformations as in real images. In our evaluation study, we aimed to measure accuracy directly in clinical longitudinal breast MR images. Approaches based on ROIs have been used more often for well-defined and highly localized structures such as some cortical structures in the brain, whereas the human breast does not necessarily have many such well-defined structures across individuals. For these reasons, registration accuracy in our evaluation study was assessed based on expert-defined landmarks.
As conceptually depicted in Figure 3, we measured interexpert landmark errors as well as algorithm-to-expert landmark errors. Assume that we have a set of expert-annotated landmarks in the baseline image (N is 85 on average, as section Dataset and Table 1 show). Two experts (SW and EC) independently found their corresponding points in the follow-up image, denoted as and . Conversely, the deformation calculated by a registration algorithm maps the same set of landmarks to locations . We could, therefore, define interexpert landmark errors = (i.e., the length of the solid line in Fig. 3) as
| [1] |
and algorithm-to-expert landmark errors (i.e., the average length of the dashed lines in Fig. 3) as
| [2] |
where d(·, ·) is the Euclidean distance between two voxel locations.
FIG. 3.
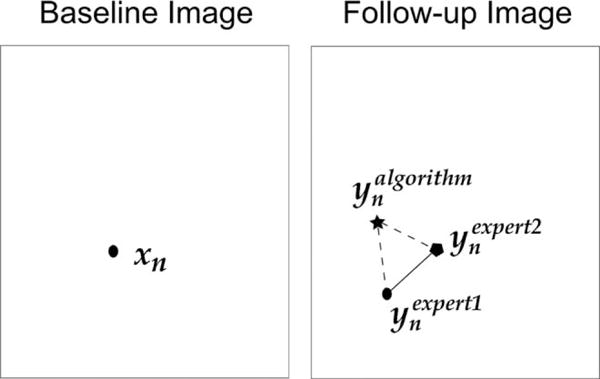
Depiction of the interexpert landmark errors (the length of the solid line) and the algorithm-to-expert landmark errors (the average length of the dashed lines).
We can further analyze landmark errors for different regions, based on the prior knowledge whether a landmark xn belongs to tumor or normal tissue regions. We can also analyze landmark errors in patient subgroups having different responses to neoadjuvant chemotherapy, based on the prior knowledge of whether a patient belongs to the pCR or pPR subgroup.
Using Registration to Quantify Tumor Changes During Neoadjuvant Chemotherapy
The registration method obtaining the highest accuracy (i.e., smallest landmark errors) was used to quantify tumor changes in patients undergoing neoadjuvant chemotherapy. We first registered the follow-up images of each patient to their baseline scans. The obtained deformation fields were further used to calculate the Jacobian determinant maps. The Jacobian determinants, computed at the voxel level, show the volumetric change ratio (follow-up/baseline) at each voxel. Mathematically, suppose a 3D baseline image is indexed by three orthogonal axes i; j; k in a Cartesian coordinate system, a voxel is denoted as u = (ui; uj; uk), and a deformation field computed from the registration of follow-up to baseline images is denoted as h(u) = (hi(u), hj(u), hk(u)) at every voxel u. Then the Jacobian determinant at voxel u, denoted as JacDet(u), is defined as
| [3] |
where
| [4] |
and det (·) is the determinant of a matrix. The Jacobian determinant value, therefore, shows the expansion of a voxel if greater than 1, shrinkage if smaller than 1, and volume-preservation if exactly 1.
RESULTS
Evaluation Results and Observations
This section presents our evaluation results. We measured the landmark-based registration accuracy in the entire image, in different regions (tumor vs. normal tissue), and in different patient subgroups having different pathologic responses to chemotherapy. The results are presented in the next three subsections, respectively. Note that we stratified registration accuracy by regions and pathologic subgroups, since it is our hypothesis that patients in the pCR subgroup may pose higher levels of difficulty to registration due to potentially larger changes in the tumor regions after treatment.
Overall Landmark Errors
The mean and standard deviation errors for different registration methods are listed in Table 2. Furthermore, Figure 4 shows the box-whisker plot of the landmark errors. Several observations can be made:
The average landmark error between human experts was 3.12 ± 2.84 mm. This error was higher than the errors in the registration of short-interval longitudinal breast MR images (usually pre- and postcontrast DCE-MRI), where studies have reported subvoxel registration accuracy (50,52). This showed the increased level of registration difficulty in longitudinal breast MR images acquired days or weeks apart. It was perhaps caused by several factors, including the patient repositioning, the soft tissue deformation, and the tumor changes due to treatment.
The landmark errors of all the automated registration methods evaluated were larger than the average error between human experts.
Among the automated methods included in our evaluation, DRAMMS performed closest to human experts, with landmark errors at 6.05 ± 4.86 mm. FFD and its variants followed, in the order of CC-FFD, NMI-FFD, and SSD-FFD, all with a statistically significant difference from DRAMMS. Demons and Diffeomorphic Demons had the largest landmark errors, with errors at 10.76 ± 6.01 mm and 10.82 ± 6.11 mm, respectively (which are similar to the 11.67 ± 4.81 mm error reported in (73) on a similar dataset). The difference between Demons and Diffeomorphic Demons was not significant (P = 0.0474), if we only consider the differences with P < 0.01 to be statistically significant.
Table 2.
Mean and Standard Deviation (stdev) of Landmark Errors for Different Registration Methods (mm)
| Interexpert | Demons | Diff. Demons | DRAMMS | CC-FFD | NMI-FFD | SSD-FFD | |
|---|---|---|---|---|---|---|---|
| Mean | 3.12 | 10.76 | 10.82 | 6.05 | 8.07 | 8.21 | 9.46 |
| Stdev | 2.84 | 6.01 | 6.11 | 4.86 | 3.86 | 3.81 | 4.55 |
Numbers in the bold font indicate either the landmark errors between human experts, or the smallest landmark errors (mean or standard deviation) of automated algorithms as compared with human experts.
Landmark Errors in Different Regions
We further examined landmark errors in tumor versus normal tissue regions. As Figure 5 shows, methods followed the same rank for landmark errors in tumor regions as for landmark errors in the entire image. In tumor regions, DRAMMS had the smallest landmark errors among all automated methods (5.64 ± 4.79 mm), then NMI/CC/SSD-FFD (8.33 ± 3.87, 8.33 ± 3.88, and 8.74 ± 4.38 mm), and then Demons (9.28 ± 5.42 mm) and Diffeomorphic Demons (9.45 ± 5.57 mm). In normal tissue regions, DRAMMS had the smallest landmark errors among all automated methods (6.41 ± 4.90 mm), then CC/NMI/SSD-FFD (8.11 ± 3.76 mm, 7.87 δ3.83 mm, 9.29 ± 4.68 mm) and then Diffeomorphic Demons (12.07 ± 6.32 mm) and Demons (12.09 ± 6.22 mm).
FIG. 5.
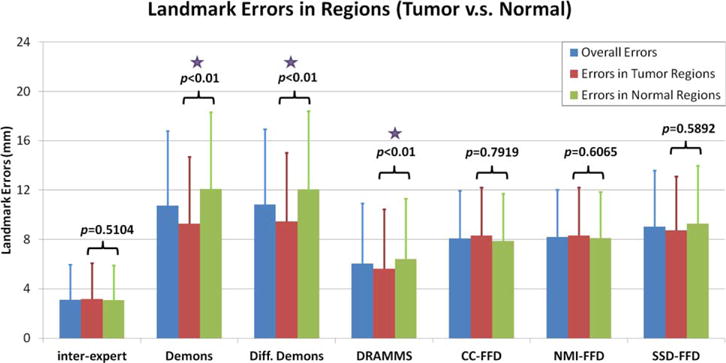
Landmark errors in different image regions. The P-values from Student’s two-sample t-test represent the significance of difference between landmark errors in the tumor versus in the normal tissue regions. Purple stars note registration methods which obtained significantly lower errors in the tumor regions compared to errors in the normal tissue regions.
One noteworthy finding is that, DRAMMS, Demons, and Diffeomorphic Demons all had significantly smaller landmark errors in the tumor regions than in the normal tissue regions (evidenced by the P-values provided in Fig. 5). One possible explanation may be that, the soft normal breast tissues exhibit more complicated deformations than the relatively solid tumors. This might be the case especially when considering factors such as patient movements and breast positioning differences in different visits, which may affect the appearances of normal tissue regions more than the tumor regions. Another potential explanation might be that the heterogeneity within the tumor regions may have helped the automated registration algorithms to better locate anatomical correspondences. Conversely, as observed in Figure 6b,c, the errors in tumor regions were actually larger than in normal tissue regions for the pCR subgroup. This suggested that the effect of the treatment may override the comparison of landmark errors in tumor versus normal tissue regions. Finally, whether the landmark differences were significant in different regions seems to also depend on which registration algorithm was used (see Figs 5 and 6). Considering all these possible factors, a larger-scale, carefully controlled study may be needed to fully evaluate the reason why and how landmark errors may differ in tumor versus normal tissue regions.
FIG. 6.
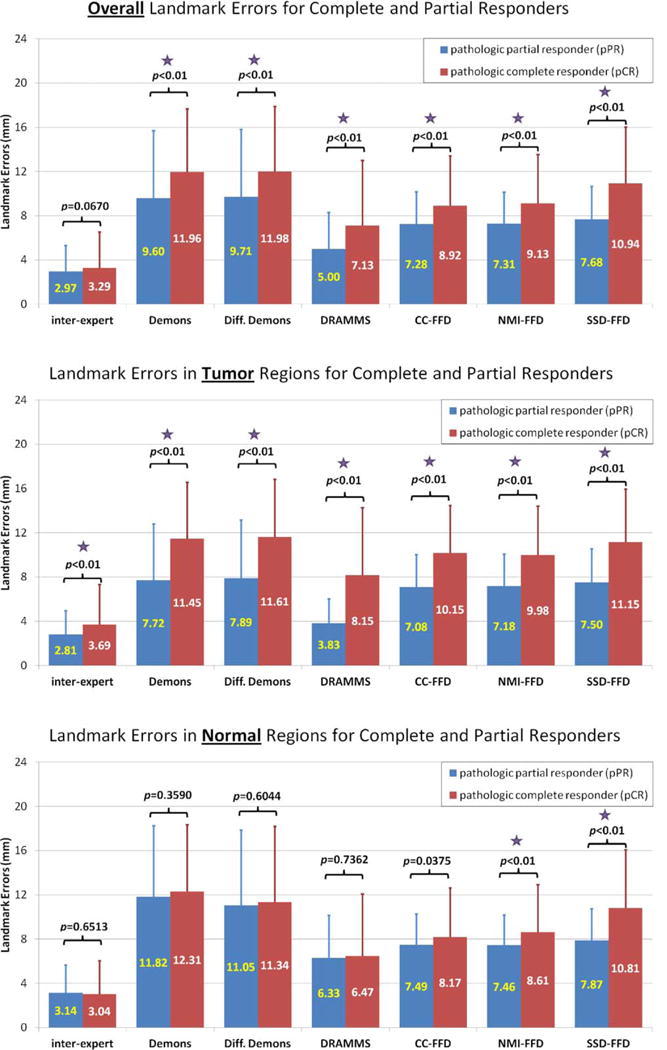
Comparison of landmark errors in the pCR and pPR patient subgroups, in (a) the entire image; (b) the tumor regions; and (c) the normal regions. The differences with P-values smaller than 0.01 are treated as statistically significant, which are noted by the purple stars in the figure.
Landmark Errors and Response to Chemotherapy
The previous subsection showed that registration accuracy may vary by image regions. This subsection will show that registration accuracy may also vary by patient subgroups. Figure 6 compares the landmark errors between the pCR and pPR subgroups, in (a) the entire image, (b) tumor regions, and (c) normal tissue regions. The P-values from the Student’s t-test were provided and the statistical significance (when P < 0.01) was noted by purple stars in the figure. Some observations can be made:
The pPR patients almost always had smaller registration errors than pCR patients, regardless of the registration methods being used, and regardless of tumor versus normal tissue regions.
The difference of the registration errors between the pPR and pCR patient subgroups were almost always statistically significant in the entire image, except for the expert raters (upper subfigure in Fig. 6). This was most likely driven by the statistically significant difference in landmark errors between the pPR and pCR patients in the tumor regions, except for the expert raters (middle subfigure). Conversely, registration errors in the normal tissue regions remained almost the same, regardless of a patient’s response to neoadjuvant chemotherapy.
Combining these two observations, the larger registration errors in the pCR subgroup as compared with the pPR subgroup may be partly because the tumors changed more in the pCR subgroup than in the pPR subgroup, which posed greater challenges especially for the automated registration methods.
Application to the Quantification of Tumor Changes
Because DRAMMS had higher accuracy than other automated registration methods evaluated, in this section, we show preliminary experiments of using the DRAMMS registration algorithm to quantify tumor changes in patients undergoing neoadjuvant chemotherapy. Examples of the registered images are shown in the second rows of Figures 7 and 8, for a representative pPR patient and a representative pCR patient, respectively. Jacobian determinant maps were computed from the obtained deformation fields, as shown in the third rows in Figures 7 and 8, which are overlaid onto the baseline intensity images. Here, the Jacobian determinant maps are shown only within tumor regions. The tumor regions were obtained in the baseline images by a user-interactive intensity thresholding step, followed by a human expert’s verification of the segmented tumor boundary. Note that the tumor regions were in no means used for image registration or for the calculation of Jacobian determinant maps; they were only used as masks to visualize the Jacobian determinants specifically within the tumor regions. As shown in Figures 7 and 8, the quantified volumetric changes agree with the visual findings of how tumors have changed due to the effect of treatment. Moreover, the Jacobian determinant maps can show the spatially heterogeneous changes within the tumor regions, which may be important features for predicting the long-term pathologic response as suggested in recent studies (30–33). Our future work will be to further validate the automatically computed heterogeneous tumor changes as quantitative imaging biomarkers for predicting the response to neoadjuvant chemotherapy.
FIG. 8.
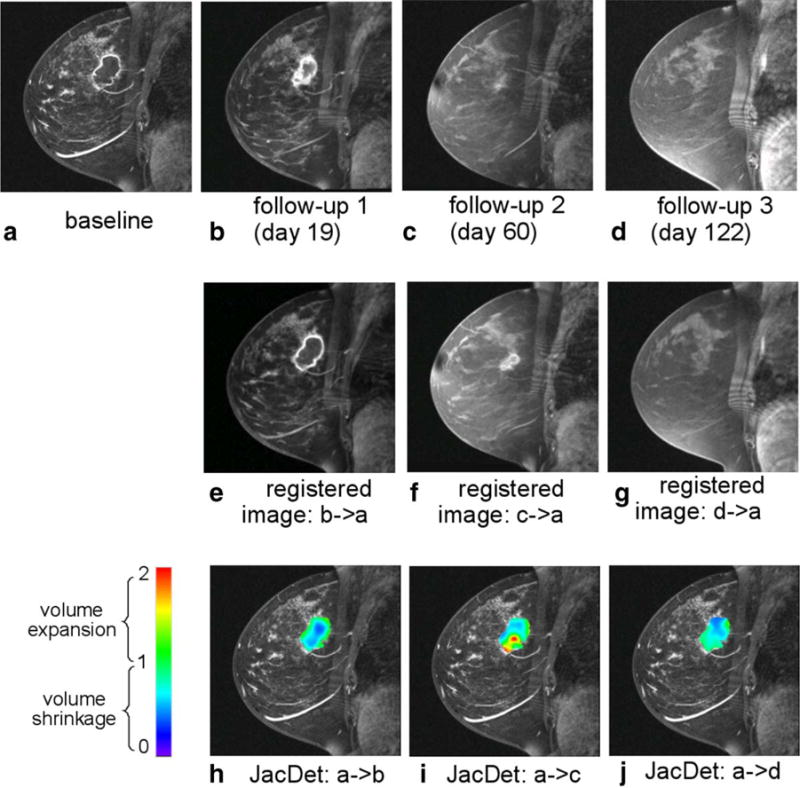
Quantification of the voxelwise longitudinal breast cancer change by the DRAMMS registration algorithm. Results shown in this figure are from an example pathologic complete responder (pCR) patient. Jacobian determinants in the tumor regions are overlaid on the baseline image in the bottom row.
DISCUSSION
Summary of Work and Findings
Deformable image registration can automatically capture 3-D volumetric changes at the voxel level. It is, therefore, an appropriate choice for quantifying the spatially heterogeneous changes within tumors. Our study evaluated DRAMMS, an attribute-based image registration method, in comparison to five other intensity-based algorithms, for the task of registering long-interval longitudinal breast MR images. We compared the accuracies of registration methods using expert-defined landmarks as references. We also showed preliminary results of using the DRAMMS registration method, which had the smallest landmark errors, to quantify longitudinal tumor changes induced by treatment. As such, our work adds into the literature several important findings:
First, automated registration methods differed greatly in their performances. The attribute-matching component helped DRAMMS to obtain the smallest landmark errors, almost 25–40% smaller than the errors from five other intensity-based methods. The high-dimensional geometric/texture attributes that DRAMMS uses seemed to be less sensitive to the image noise, the intensity inhomogeneity, or the partial volume effects. Because of this, DRAMMS also seemed to require fewer parameter tunings. Besides longitudinal breast images presented in this study, another recent study on the intersubject brain image registration also found a similar trend, that DRAMMS performed relatively accurately, without much need for parameter tunings (80).
Second, our analysis on image regions and patient subgroups showed that, the voxel correspondences were least ambiguous in the tumor regions of patients in the pPR subgroup (where tumors expanded or shrank but did not disappear), then a little more ambiguous in the normal tissue regions (almost equivalent between the pCR and pPR subgroups), and most ambiguous in the tumor regions of patients in the pCR subgroup (perhaps because tumors changed more, or even disappeared, which caused a possible loss of correspondences in some locations). This trend has been observed for both experts and most algorithms, as can be observed from the landmark errors in panels (b) and (c) of Figure 6.
Third, because of the higher accuracy of DRAMMS compared to other registration methods, we further evaluated its ability to quantify longitudinal tumor changes as induced by neoadjuvant chemotherapy for breast cancer. Although preliminary, our results were promising. Those results showed the ability of DRAMMS to capture the changes agreeable to human observations. Furthermore, those results also showed the ability of DRAMMS to characterize the spatially heterogeneous effects induced by treatment, which may be ultimately used as more sensitive markers for evaluating treatment response and predicting patient outcomes.
Future Work
On further evaluating the performance of registering longitudinal breast MR images, our future work will include (a) the quantitative evaluation of registration accuracies on longitudinal images with more than one follow-up time point; (b) the evaluation of the performance of registration in larger datasets and for a broader range of registration algorithms [although not being reported before as being used in this context, many deformable registration algorithms/tools may well serve this specific registration task, such as elastix (81), ANTs (82), ART (83), DROP (76), HAMMER (84), MIND (85), NiftyReg (86), and plastimath (87)]; and (c) the evaluation of the change of registration performance as registration parameters vary. Also, the quantification and better understanding of the genuine ambiguity for human experts to annotate landmarks is another important topic to explore. It may require a substantial amount of work, as it could involve many factors such as varying landmark locations and deriving some precise quantitative measures of the intra-rater ambiguity.
On further improving the accuracy of registering longitudinal breast MR images, our future work will extend the current 3-D registration into 4-D (3-D + time) or groupwise registration [e.g., (54,88–92)]. As tumor changes are often temporally smooth, registering a full 4-D spectrum of longitudinal images at the same time can better maintain the temporal smoothness. In addition, we can incorporate some task-specific prior knowledge to further aid the registration, such as the removal of image background and the chest region [e.g., (93–95)], the automatically detected landmarks [e.g., (96,97)], the segmentation of various breast tissue types [e.g., tumor vs. normal fibroglandular tissue (98,99)], and even the explicit segmentation of tumors [e.g., (100–102)]. Such modifications could further aid the registration to focus on ROI, and could largely reduce the negative impacts from structures that are not of interest to the specific application. Finally, we could also explore methodologies to form a consensus mechanism, where multiple registration methods can complement each other toward more accurately capturing voxelwise volumetric changes [e.g., (103,104)].
Finally, based on our reported results, our future work will also directly evaluate registration-quantified tumor changes. This would include (a) a direct comparison with the RECIST and other segmentation-based approaches, for the accuracy of the quantified morphologic changes; and (b) the use of registration-quantified changes and the registration-revealed heterogeneity of changes to predict long-term pathologic response to treatment. This should have an impact on treatment optimization for breast cancer patients.
CONCLUSION
The registration of longitudinal breast MR images acquired days or weeks apart during the course of treatment has gained increasing interest as a tool, because it can be used to quantify tumor changes, assess treatment effects, and potentially predict long-term response and patient survival. This article performed a comprehensive evaluation of the registration of longitudinal breast MR image, using expert-annotated landmarks as references. Specifically, we evaluated DRAMMS, an attribute-based registration method, and compared it with five other intensity-based registration methods. Our results showed that DRAMMS can achieve significant improvements in registration accuracy over other automated registration methods included in this study. This was possibly due to the use of the high-dimensional voxel neighborhood information to characterize the geometric and anatomical properties of voxels, which seemed to be more robust to the noise, the intensity inhomogeneity and the contrast changes between images. We also showed some preliminary evidence of using DRAMMS to quantify the longitudinal tumor changes, and the spatial heterogeneity, as induced by neoadjuvant chemotherapy. Our ultimate goal is to use registration-quantified tumor changes to more accurately predict long-term pathologic response and patient survival after treatment.
Acknowledgments
The authors would like to thank Dr. Nola Hylton from UCSF for the permission to use the I-SPY data and the useful discussions on this research work.
Grant sponsor: pilot grant from the University of Pennsylvania Center for Biomedical Image Computing and Analytics (CBICA).
References
- 1.American Cancer Society. Cancer facts and figures. 2013 Available at: http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-036845.pdf. 2013. Accessed on November 16, 2013.
- 2.Hylton NM, Blume JD, Bernreuter WK, et al. Locally advanced breast cancer: MR imaging for prediction of response to neoadjuvant chemotherapy results from ACRIN 6657/i-spy trial. Radiology. 2012;263:663–672. doi: 10.1148/radiol.12110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall FM. MR imaging for the prediction of breast cancer response to neoadjuvant chemotherapy. Radiology. 2013;266:367–367. doi: 10.1148/radiol.12124051. [DOI] [PubMed] [Google Scholar]
- 4.Bonadonna G, Valagussa P, Brambilla C, Ferrari L, Moliterni A, Terenziani M, Zambetti M. Primary chemotherapy in operable breast cancer: eight-year experience at the milan cancer institute. J Clin Oncol. 1998;16:93–100. doi: 10.1200/JCO.1998.16.1.93. [DOI] [PubMed] [Google Scholar]
- 5.Fisher B, Bryant J, Wolmark N, et al. Effect of preoperative chemotherapy on the outcome of women with operable breast cancer. J Clin Oncol. 1998;16:2672–2685. doi: 10.1200/JCO.1998.16.8.2672. [DOI] [PubMed] [Google Scholar]
- 6.Baek H, Chen J, Nie K, Hon J, Bahri S, Mehta R, Nalcioglu O, Su M. Predicting pathologic response to neoadjuvant chemotherapy in breast cancer by using MR imaging and quantitative 1H MR spectroscopy. Radiology. 2009;251:653–662. doi: 10.1148/radiol.2512080553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLaughlin R, Hylton N. MRI in breast cancer therapy monitoring. NMR Biomed. 2011;24:712–720. doi: 10.1002/nbm.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takada M, Sugimoto M, Ohno S, et al. Predictions of the pathological response to neoadjuvant chemotherapy in patients with primary breast cancer using a data mining technique. Breast Cancer Res Treat. 2012;134:661–670. doi: 10.1007/s10549-012-2109-2. [DOI] [PubMed] [Google Scholar]
- 9.Abramson RG, Li X, Hoyt TL, Su PF, Arlinghaus LR, Wilson KJ, Abramson VG, Chakravarthy AB, Yankeelov TE. Early assessment of breast cancer response to neoadjuvant chemotherapy by semi-quantitative analysis of high-temporal resolution DCE-MRI: preliminary results. Magn Reson Imaging. 2013;31:1457–1464. doi: 10.1016/j.mri.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weis JA, Miga MI, Arlinghaus LR, Li X, Chakravarthy AB, Abramson V, Farley J, Yankeelov TE. A mechanically coupled reaction–diffusion model for predicting the response of breast tumors to neoadjuvant chemotherapy. Phys Med Biol. 2013;58:5851–5866. doi: 10.1088/0031-9155/58/17/5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abramson RG, Arlinghaus LR, Weis JA, Li X, Dula AN, Chekmenev EY, Smith SA, Miga MI, Abramson VG, Yankeelov TE. Current and emerging quantitative magnetic resonance imaging methods for assessing and predicting the response of breast cancer to neoadjuvant therapy. Breast Cancer. 2012;2012:139–154. doi: 10.2147/BCTT.S35882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atuegwu NC, Arlinghaus LR, Li X, Chakravarthy AB, Abramson VG, Sanders ME, Yankeelov TE. Parameterizing the logistic model of tumor growth by DW-MRI and DCE-MRI data to predict treatment response and changes in breast cancer cellularity during neoadjuvant chemotherapy. Transl Oncol. 2013;6:256–264. doi: 10.1593/tlo.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Arlinghaus LR, Ayers GD, et al. DCE-MRI analysis methods for predicting the response of breast cancer to neoadjuvant chemotherapy: pilot study findings. Magn Reson Med. 2014;71:1592–1602. doi: 10.1002/mrm.24782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mani S, Chen Y, Li X, Arlinghaus L, Chakravarthy AB, Abramson V, Bhave SR, Levy MA, Xu H, Yankeelov TE. Machine learning for predicting the response of breast cancer to neoadjuvant chemotherapy. J Am Med Inform Assoc. 2013;20:688–695. doi: 10.1136/amiajnl-2012-001332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajan R, Esteva F, Symmans W. Pathologic changes in breast cancer following neoadjuvant chemotherapy: implications for the assessment of response. Clin Breast Cancer. 2004;5:235–238. doi: 10.3816/cbc.2004.n.028. [DOI] [PubMed] [Google Scholar]
- 16.Tateishi U, Miyake M, Nagaoka T, Terauchi T, Kubota K, Kinoshita T, Daisaki H, Macapinlac H. Neoadjuvant chemotherapy in breast cancer: prediction of pathologic response with PET/CT and dynamic contrast-enhanced MR imaging—prospective assessment. Radiology. 2012;263:53–63. doi: 10.1148/radiol.12111177. [DOI] [PubMed] [Google Scholar]
- 17.Lin M, Chen J, Mehta R, Bahri S, Chan S, Nalcioglu O, Su M. Spatial shrinkage/expansion patterns between breast density measured in two MRI scans evaluated by non-rigid registration. Phys Med Biol. 2011;56:5865–5875. doi: 10.1088/0031-9155/56/18/006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar S, Johnson T, Ma B, Chenevert T, Bland P, Park H, Schott A, Ross B, Meyer C. Evaluation of an automatic registration-based algorithm for direct measurement of volume change in tumors. Int J Radiat Oncol Biol Phys. 2012;83:1038–1046. doi: 10.1016/j.ijrobp.2011.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ou Y, Sotiras A, Paragios N, Davatzikos C. DRAMMS: deformable registration via attribute matching and mutual-saliency weighting. Med Image Anal. 2011;15:622–639. doi: 10.1016/j.media.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Londero V, Bazzocchi M, Del Frate C, Puglisi F, Di Loreto C, Francescutti G, Zuiani C. Locally advanced breast cancer: comparison of mammography, sonography and MR imaging in evaluation of residual disease in women receiving neoadjuvant chemotherapy. Eur Radiol. 2004;14:1371–1379. doi: 10.1007/s00330-004-2246-z. [DOI] [PubMed] [Google Scholar]
- 21.Wasif N, Garreau J, Terando A, Kirsch D, Mund D, Giuliano A. MRI versus ultrasonography and mammography for preoperative assessment of breast cancer. Am Surg. 2009;75:970–975. [PubMed] [Google Scholar]
- 22.Partridge S, Gibbs J, Lu Y, Esserman L, Tripathy D, Wolverton D, Rugo H, Hwang E, Ewing C, Hylton N. MRI measurements of breast tumor volume predict response to neoadjuvant chemotherapy and recurrence-free survival. Am J Roentgenol. 2005;184:1774–1781. doi: 10.2214/ajr.184.6.01841774. [DOI] [PubMed] [Google Scholar]
- 23.Nie K, Chen J, Chan S, Chau M, Yu H, Bahri S, Tseng T, Nalcioglu O, Su M. Development of a quantitative method for analysis of breast density based on three-dimensional breast MRI. Med Phys. 2008;35:5253–5262. doi: 10.1118/1.3002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Symmans W, Liu J, Knowles D, Inghirami G. Breast cancer heterogeneity: evaluation of clonality in primary and metastatic lesions. Human Pathol. 1995;26:210–216. doi: 10.1016/0046-8177(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 25.Degani H, Gusis V, Weinstein D, Fields S, Strano S. Mapping pathophysiological features of breast tumors by MRI at high spatial resolution. Nat Med. 1997;3:780–782. doi: 10.1038/nm0797-780. [DOI] [PubMed] [Google Scholar]
- 26.Sharifi-Salamatian V, Pesquet-Popescu B, Simony-Lafontaine J, Rigaut J. Index for spatial heterogeneity in breast cancer. J Microsc. 2004;216:110–122. doi: 10.1111/j.0022-2720.2004.01398.x. [DOI] [PubMed] [Google Scholar]
- 27.Anderson W, Matsuno R. Breast cancer heterogeneity: a mixture of at least two main types? J Natl Cancer Inst. 2006;98:948–951. doi: 10.1093/jnci/djj295. [DOI] [PubMed] [Google Scholar]
- 28.Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- 29.Polyak K. Heterogeneity in breast cancer. J Clin Investig. 2011;121:3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maday P, Khurd P, Ladic L, Schnall M, Rosen M, Davatzikos C, Kamen A. Imaging as a surrogate for the early prediction and assessment of treatment response through the analysis of 4-d texture ensembles (ISEPARATE) Med Comput Vis Recogn Tech Appl Med Imaging. 2011:164–173. [Google Scholar]
- 31.Li X, Arlinghaus L, Chakravarthy A, Farley J, Mayer I, Abramson V, Kelley M, Meszoely I, Means-Powell J, Yankeelov T. Workshop on Biomedical Image Registration (WBIR) Nashville, Tennessee, USA: 2012. Early DCE-MRI changes after longitudinal registration may predict breast cancer response to neoadjuvant chemotherapy; pp. 229–235. [Google Scholar]
- 32.Li X, Arlinghaus L, Chakravarthy A, et al. Voxel-based analysis of early DCE-MRI changes may predict the response to neoadjuvant chemotherapy in breast cancer patients. Proceedings of the 20th Annual Meeting of ISMRM; Melbourne, Australia. 2012. p. 1465. [Google Scholar]
- 33.De Los Santos JF, Cantor A, Amos KD, et al. Magnetic resonance imaging as a predictor of pathologic response in patients treated with neoadjuvant systemic treatment for operable breast cancer. Cancer. 2013;119:1776–1783. doi: 10.1002/cncr.27995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones EF, Sinha SP, Newitt DC, Klifa C, Kornak J, Park CC, Hylton NM. MRI enhancement in stromal tissue surrounding breast tumors: association with recurrence free survival following neoadjuvant chemotherapy. PloS One. 2013;8:e61969. doi: 10.1371/journal.pone.0061969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein A, Andersson J, Ardekani B, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46:786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ants similarity metric performance in brain image registration. NeuroImage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy K, van Ginneken B, Reinhardt J, Kabus S, Ding K, Deng X, Pluim J. Evaluation of methods for pulmonary image registration: the EMPIRE10 study. IEEE Trans Med Imaging. 2011;30:1901–1920. doi: 10.1109/TMI.2011.2158349. [DOI] [PubMed] [Google Scholar]
- 38.Ou Y, Ye DH, Pohl KM, Davatzikos C. Biomedical image registration. Springer; 2012. Validation of DRAMMS among 12 popular methods in cross-subject cardiac MRI registration; pp. 209–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dura E, Domingo J, Ayala G, Mart ı-Bonmat ı L. Evaluation of the registration of temporal series of contrast-enhanced perfusion magnetic resonance 3d images of the liver. Comput Methods Programs Biomed. 2012;108:932–945. doi: 10.1016/j.cmpb.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Cifor A, Risser L, Chung D, Anderson E, Schnabel J. Hybrid feature-based diffeomorphic registration for tumour tracking in 2-d liver ultrasound images. IEEE Trans Med Imaging. 2013;32:1647–1656. doi: 10.1109/TMI.2013.2262055. [DOI] [PubMed] [Google Scholar]
- 41.Bakic P, Albert M, Brzakovic D, Maidment A. Mammogram synthesis using a 3d simulation. i. Breast tissue model and image acquisition simulation. Med Phys. 2002;29:2131. doi: 10.1118/1.1501143. [DOI] [PubMed] [Google Scholar]
- 42.van Engeland S, Snoeren P, Hendriks J, Karssemeijer N. A comparison of methods for mammogram registration. IEEE Trans Med Imaging. 2003;22:1436–1444. doi: 10.1109/TMI.2003.819273. [DOI] [PubMed] [Google Scholar]
- 43.Filev P, Hadjiiski L, Sahiner B, Chan H, Helvie M. Comparison of similarity measures for the task of template matching of masses on serial mammograms. Med Phys. 2005;32:515. doi: 10.1118/1.1851892. [DOI] [PubMed] [Google Scholar]
- 44.Richard F, Bakic P, Maidment A. Mammogram registration: a phantom-based evaluation of compressed breast thickness variation effects. IEEE Trans Med Imaging. 2006;25:188–197. doi: 10.1109/TMI.2005.862204. [DOI] [PubMed] [Google Scholar]
- 45.Hipwell J, Tanner C, Crum W, Schnabel JA, Hawkes D. A new validation method for X-ray mammogram registration algorithms using a projection model of breast X-ray compression. IEEE Trans Med Imaging. 2007;26:1190–1200. doi: 10.1109/TMI.2007.903569. [DOI] [PubMed] [Google Scholar]
- 46.Diez Y, Oliver A, Llado X, Marti R. Comparison of registration methods using mamographic images In 17th. IEEE International Conference on Image Processing (ICIP), IEEE; Hongkong, China. 2010. pp. 4421–4424. [Google Scholar]
- 47.Pereira S, Hipwell J, McCormack V, et al. Automated registration of diagnostic to prediagnostic X-ray mammograms: evaluation and comparison to radiologists accuracy. Med Phys. 2010;37:4530–4539. doi: 10.1118/1.3457470. [DOI] [PubMed] [Google Scholar]
- 48.Denton E, Sonoda L, Rueckert D, Rankin S, Hayes C, Leach M, Hill D, Hawkes D. Comparison and evaluation of rigid, affine, and nonrigid registration of breast MR images. J Comput Assist Tomogr. 1999;23:800–805. doi: 10.1097/00004728-199909000-00031. [DOI] [PubMed] [Google Scholar]
- 49.Tanner C, Schnabel J, Degenhard A, Castellano-Smith A, Hayes C, Leach M, Hose D, Hill D, Hawkes D. Medical Image Computing and Computer-Assisted Intervention–MICCAI. Tokyo, Japan: 2002. Validation of volume-preserving non-rigid registration: application to contrast-enhanced MR-mammography; pp. 307–314. [Google Scholar]
- 50.Schnabel J, Tanner C, Castellano-Smith A, Degenhard A, Leach M, Hose D, Hill D, Hawkes D. Validation of nonrigid image registration using finite-element methods: application to breast MR images. IEEE Trans Med Imaging. 2003;22:238–247. doi: 10.1109/TMI.2002.808367. [DOI] [PubMed] [Google Scholar]
- 51.Tanner C, Schnabel JA, Hill D, Hawkes D, Degenhard A, Leach M, Hose D, Hall-Craggs M, Usiskin S. Quantitative evaluation of free-form deformation registration for dynamic contrast-enhanced MR mammography. Med Phys. 2007;34:1221–1233. doi: 10.1118/1.2712040. [DOI] [PubMed] [Google Scholar]
- 52.Martel A, Froh M, Brock K, Plewes D, Barber D. Evaluating an optical-flow-based registration algorithm for contrast-enhanced magnetic resonance imaging of the breast. Phys Med Biol. 2007;52:3803. doi: 10.1088/0031-9155/52/13/010. [DOI] [PubMed] [Google Scholar]
- 53.Hill A, Mehnert A, Crozier S, McMahon K. Evaluating the accuracy and impact of registration in dynamic contrast-enhanced breast MRI. Concepts Magn Reson Part B Magn Reson Eng. 2009;35:106–120. [Google Scholar]
- 54.Kim M, Wu G, Shen D. Hierarchical alignment of breast DCE-MR images by groupwise registration and robust feature matching. Med Phys. 2012;39:353–366. doi: 10.1118/1.3665705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuczynski K, Siczek M, Stegierski R. Application of image registration techniques in dynamic magnetic resonance imaging of breast. J Med Inform. 2011;17:275–280. [Google Scholar]
- 56.Rohlfing T, Maurer C, Jr, Bluemke D, Jacobs M. Volume-preserving nonrigid registration of MR breast images using free-form deformation with an incompressibility constraint. IEEE Trans Med Imaging. 2003;22:730–741. doi: 10.1109/TMI.2003.814791. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, Dang J, Du X, Li S. A non-rigid registration method for dynamic contrast enhancement breast MRI. Intell Comput Inform Sci. 2011:644–649. [Google Scholar]
- 58.Crum W, Tanner C, Hawkes D. Anisotropic multi-scale fluid registration: evaluation in magnetic resonance breast imaging. Phys Med Biol. 2005;50:5153–5174. doi: 10.1088/0031-9155/50/21/014. [DOI] [PubMed] [Google Scholar]
- 59.Hayton P, Brady M, Smith S, Moore N. A non-rigid registration algorithm for dynamic breast MR images. Artif Intell. 1999;114:125–156. [Google Scholar]
- 60.Tanner C, Schnabel J, Chung D, Clarkson M, Rueckert D, Hill D, Hawkes D. Medical Image Computing and Computer-Assisted Intervention–MICCAI. Pittsburgh, PA: Springer; 2000. Volume and shape preservation of enhancing lesions when applying non-rigid registration to a time series of contrast enhancing MR breast images; pp. 327–337. [Google Scholar]
- 61.Arlinghaus L, Li X, Levy M, Smith D, Welch E, Gore J, Yankeelov T. Current and future trends in magnetic resonance imaging assessments of the response of breast tumors to neoadjuvant chemotherapy. J Oncol. 2010:1–17. doi: 10.1155/2010/919620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roose L, Loeckx D, Mollemans W, Maes F, Suetens P. Medical Image Computing and Computer-Assisted Intervention–MICCAI. New York City, NY, USA: 2008. Adaptive boundary conditions for physically based follow-up breast MR image registration; pp. 839–846. [DOI] [PubMed] [Google Scholar]
- 63.Esserman L, Perou C, Cheang M, DeMichele A, Carey L, van’t Veer L, Gray J, Petricoin E, Conway K, Berry D. Breast cancer molecular profiles and tumor response of neoadjuvant doxorubicin and paclitaxel: the i-spy trial (calgb 150007/150012, acrin 6657) J Clin Oncol. 2009;27(18 Suppl):515. [Google Scholar]
- 64.Barker A, Sigman C, Kelloff G, Hylton N, Berry D, Esserman L. I-spy 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin Pharmacol Ther. 2009;86:97–100. doi: 10.1038/clpt.2009.68. [DOI] [PubMed] [Google Scholar]
- 65.Chen J, Feig B, Agrawal G, Yu H, Carpenter P, Mehta R, Nalcioglu O, Su M. MRI evaluation of pathologically complete response and residual tumors in breast cancer after neoadjuvant chemotherapy. Cancer. 2008;112:17–26. doi: 10.1002/cncr.23130. [DOI] [PubMed] [Google Scholar]
- 66.von Minckwitz G, Untch M, Blohmer J, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 67.McAuliffe MJ, Lalonde FM, McGarry D, Gandler W, Csaky K, Trus BL. Medical image processing, analysis and visualization in clinical research. 14th IEEE Symposium on Computer-Based Medical Systems (CBMS 2001). Proceedings. IEEE; Bethesda, Maryland, USA. 2001. pp. 381–386. [Google Scholar]
- 68.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 69.Thirion J. Image matching as a diffusion process: an analogy with maxwell’s demons. Med Image Anal. 1998;2:243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- 70.Vercauteren T, Pennec X, Perchant A, Ayache N. Diffeomorphic demons: efficient non-parametric image registration. NeuroImage. 2008;45:S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 71.Rueckert D, Sonoda L, Hayes C, Hill D, Leach M, Hawkes D. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 72.Modersitzki J. Numerical methods for image registration, numerical mathematics and scientific computation. Oxford, UK: Oxford University Press; 2004. [Google Scholar]
- 73.Boehler T, Schilling K, Bick U, Hahn H. Workshop of Biomedical Image Registration (WBIR) Lübeck, Germany: 2010. Deformable image registration of follow-up breast magnetic resonance images; pp. 13–24. [Google Scholar]
- 74.Meyer C, Boes J, Kim B, Bland P, Zasadny K, Kison P, Koral K, Frey K, Wahl R. Demonstration of accuracy and clinical versatility of mutual information for automatic multimodality image fusion using affine and thin-plate spline warped geometric deformations. Med Image Anal. 1997;1:195–206. doi: 10.1016/s1361-8415(97)85010-4. [DOI] [PubMed] [Google Scholar]
- 75.Ou Y, Davatzikos C. Information processing in medical imaging. Springer; Williamsburgh, VA: 2009. DRAMMS: deformable registration via attribute matching and mutual-saliency weighting; pp. 50–62. [DOI] [PubMed] [Google Scholar]
- 76.Glocker B, Komodakis N, Tziritas G, Navab N, Paragios N. Dense image registration through MRFs and efficient linear programming. Med Image Anal. 2008;12:731–741. doi: 10.1016/j.media.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 77.Glocker B, Sotiras A, Komodakis N, Paragios N. Deformable medical image registration: setting the state of the art with discrete methods. Annu Rev Biomed Eng. 2011;13:219–244. doi: 10.1146/annurev-bioeng-071910-124649. [DOI] [PubMed] [Google Scholar]
- 78.Pheiffer T, Ou J, Ong R, Miga M. Automatic generation of boundary conditions using demons nonrigid image registration for use in 3-d modality-independent elastography. IEEE Trans Biomed Eng. 2011;58:2607–2616. doi: 10.1109/TBME.2011.2159791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li X, Dawant B, Welch E, Chakravarthy A, Freehardt D, Mayer I, Kelley M, Meszoely I, Gore J, Yankeelov T. A nonrigid registration algorithm for longitudinal breast MR images and the analysis of breast tumor response. Magn Reson Imaging. 2009;27:1258–1270. doi: 10.1016/j.mri.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ou Y, Akbari H, Billelo M, Da X, Davatzikos C. Comparative evaluation of registration algorithms for different brain databases with varying difficulty: Results and insights. IEEE Trans Med Imaging. 2014 doi: 10.1109/TMI.2014.2330355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 82.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ardekani BA, Braun M, Hutton BF, Kanno I, Iida H. A fully automatic multimodality image registration algorithm. J Comput Assist Tomogr. 1995;19:615–623. doi: 10.1097/00004728-199507000-00022. [DOI] [PubMed] [Google Scholar]
- 84.Shen D, Davatzikos C. HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging. 2002;21:1421–1439. doi: 10.1109/TMI.2002.803111. [DOI] [PubMed] [Google Scholar]
- 85.Heinrich MP, Jenkinson M, Bhushan M, Matin T, Gleeson FV, Brady SM, Schnabel JA. Mind: modality independent neighbourhood descriptor for multi-modal deformable registration. Med Image Anal. 2012;16:1423–1435. doi: 10.1016/j.media.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 86.Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S. Fast free-form deformation using graphics processing units. Comput Methods Programs Biomed. 2010;98:278–284. doi: 10.1016/j.cmpb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 87.Sharp G, Li R, Wolfgang J, Chen G, Peroni M, Spadea M, Mori S, Zhang J, Shackleford J, Kandasamy N. Plastimatchan open source software suite for radiotherapy image processing. Proceedings of the International Conference on the use of Computers in Radiotherapy (ICCR); Amsterdam, the Netherlands. 2010. [Google Scholar]
- 88.Shen D, Davatzikos C. Measuring temporal morphological changes robustly in brain MR images via 4-dimensional template warping. NeuroImage. 2004;21:1508–1517. doi: 10.1016/j.neuroimage.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 89.Zöllei L, Learned-Miller E, Grimson E, Wells W. Efficient population registration of 3d data In Computer Vision for Biomedical Image Applications. Beijing, China: Springer; 2005. pp. 291–301. [Google Scholar]
- 90.Geng X, Christensen GE, Gu H, Ross TJ, Yang Y. Implicit reference-based group-wise image registration and its application to structural and functional MRI. NeuroImage. 2009;47:1341–1351. doi: 10.1016/j.neuroimage.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reuter M, Rosas HD, Fischl B. Highly accurate inverse consistent registration: a robust approach. NeuroImage. 2010;53:1181–1196. doi: 10.1016/j.neuroimage.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Csapo I, Davis B, Shi Y, Sanchez M, Styner M, Neithammer M. Longitudinal image registration with temporally-dependent image similarity measure. IEEE Trans Med Imaging. 2013:1939–1951. doi: 10.1109/TMI.2013.2269814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wu S, Weinstein S, Conant E, Localio A, Schnall M, Kontos D. SPIE Medical Imaging. International Society for Optics and Photonics; San Diego, California, USA: 2012. Fully automated chest wall line segmentation in breast MRI by using context information; pp. 831507–831507. [Google Scholar]
- 94.Wu S, Weinstein S, Conant E, Schnall M, Kontos D. Automated chest wall line detection for whole-breast segmentation in sagittal breast MR images. Med Phys. 2013;40:1–12. doi: 10.1118/1.4793255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao Y, Tannenbaum A, Chen H, Torres M, Yoshida E, Yang X, Wang Y, Curran W, Liu T. Automated skin segmentation in ultrasonic evaluation of skin toxicity in breast cancer radiotherapy. Ultrasound Med Biol. 2013;39:2166–2175. doi: 10.1016/j.ultrasmedbio.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ou Y, Besbes A, Bilello M, Mansour M, Davatzikos C, Paragios N. Detecting mutually-salient landmark pairs with MRF regularization. International symposium on biomedical imaging: from nano to macro. IEEE; Rotterdam, Netherlands. 2010. pp. 400–403. [Google Scholar]
- 97.Sotiras A, Ou Y, Glocker B, Davatzikos C, Paragios N. Medical Image Computing and Computer-Assisted Intervention–MICCAI. Beijing, China: Springer; 2010. Simultaneous geometric-iconic registration; pp. 676–683. [DOI] [PubMed] [Google Scholar]
- 98.Xing Y, Ou Y, Englander S, Schnall M, Shen D. Simultaneous estimation and segmentation of t1 map for breast parenchyma measurement. 4th IEEE international symposium on biomedical imaging: from nano to macro. ISBI. IEEE; Washington DC, USA. 2007. pp. 332–335. [Google Scholar]
- 99.Wu S, Weinstein S, Kontos D. Medical Image Computing and Computer-Assisted Intervention—MICCAI. Nice, France: 2012. Atlas-based probabilistic fibroglandular tissue segmentation in breast MRI; pp. 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng Y, Englander S, Baloch S, Zacharaki E, Fan Y, Schnall M, Shen D. Step: Spatiotemporal enhancement pattern for MR-based breast tumor diagnosis. Med Phys. 2009;36:3192–3204. doi: 10.1118/1.3151811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zheng Y, Baloch S, Englander S, Schnall M, Shen D. Medical Image Computing and Computer-Assisted Intervention–MICCAI. Brisbane, Australia: 2007. Segmentation and classification of breast tumor using dynamic contrast-enhanced MR images; pp. 393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ashraf A, Gavenonis S, Daye D, Mies C, Rosen M, Kontos D. A multichannel markov random field framework for tumor segmentation with an application to classification of gene expression-based breast cancer recurrence risk. IEEE Trans Med Imaging. 2012;32:637–648. doi: 10.1109/TMI.2012.2219589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seshamani S, Rajan P, Kumar R, Girgis H, Dassopoulos T, Mullin G, Hager G. A meta registration framework for lesion matching In Medical Image Computing and Computer-Assisted Intervention– MICCAI. London: Springer; 2009. pp. 582–589. [DOI] [PubMed] [Google Scholar]
- 104.Muenzing SE, van Ginneken B, Pluim JP. Biomedical Image Registration. Nashville, TN: Springer; 2012. On combining algorithms for deformable image registration; pp. 256–265. [Google Scholar]


