Figure 3.
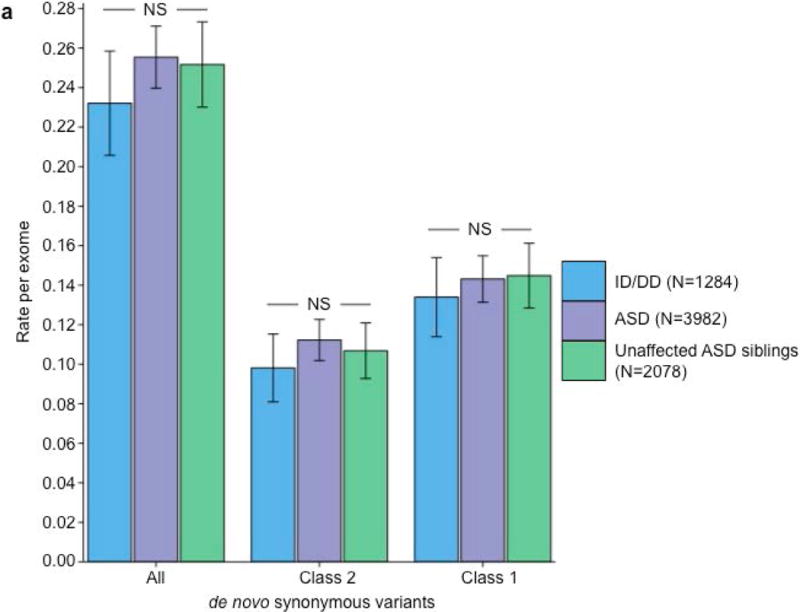
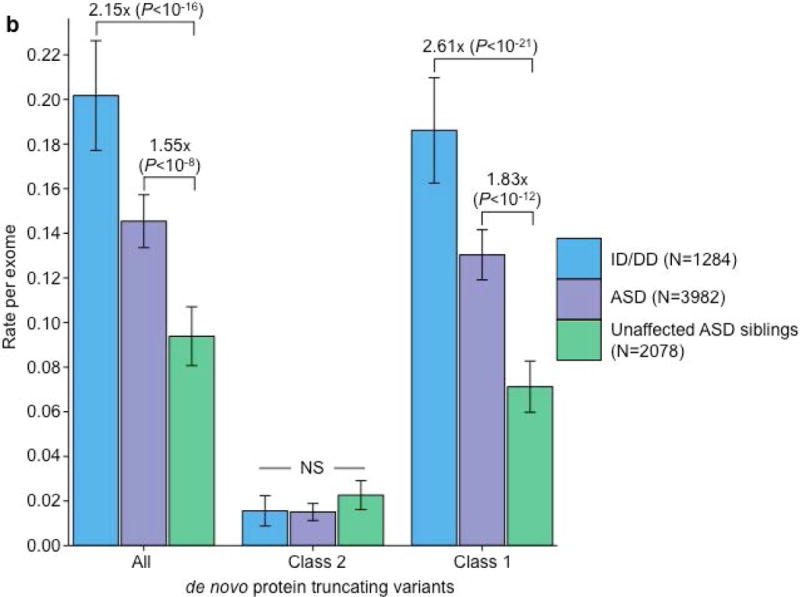
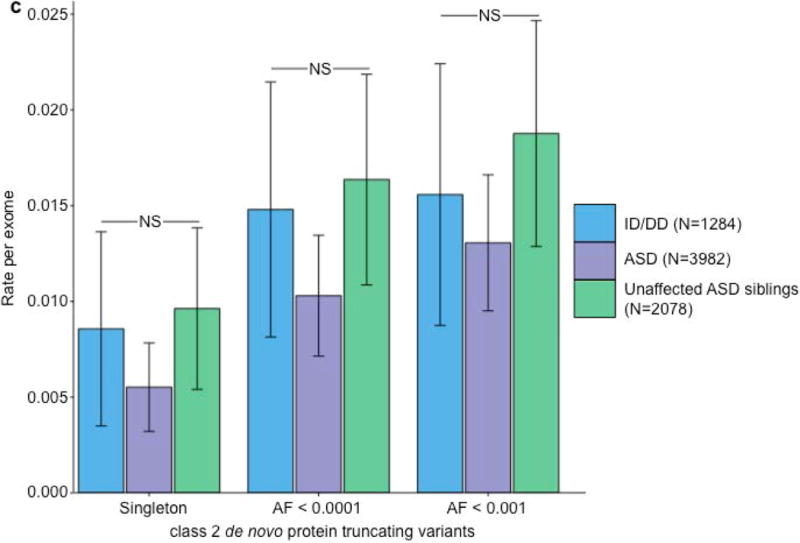
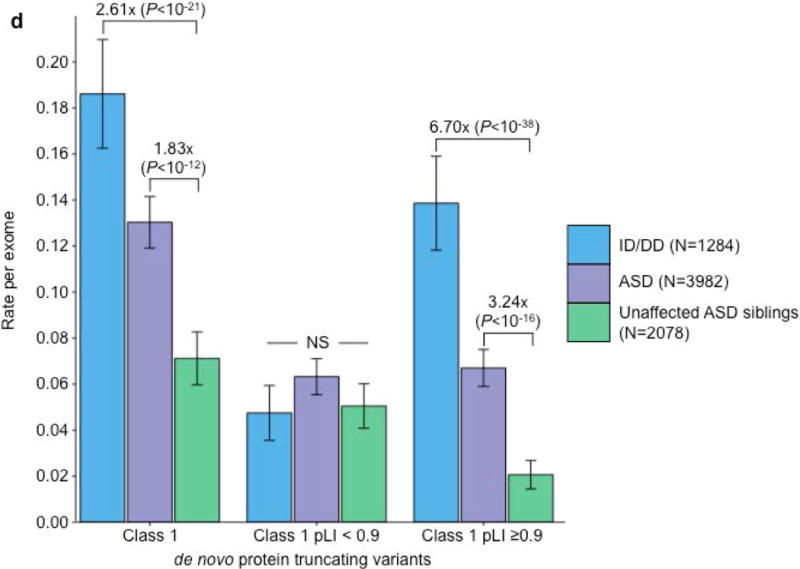
Partitioning the rate of de novo variants per exome by class 1, class 2, and pLI. Within each grouping, the rate – variants per individual – is shown for ID/DD (left), ASD (middle), and unaffected ASD siblings (right) with the number of individuals labeled in the legends. (a) Rate of de novo synonymous variants per exome partitioned into class 2 (middle) and class 1 (right). No significant difference was observed for any grouping of de novo synonymous variants. (b) Rate of de novo PTVs per exome partitioned into class 2 (middle) and class 1 (right). Only class 1 de novo PTVs in ID/DD and ASD show association when compared to unaffected ASD siblings. (c) Rate of class 2 de novo PTVs broken by different ExAC global allele frequency (AF) thresholds: singleton (observed once; left), AF < 0.0001 (middle), and AF < 0.001 (right). (d) Rate of class 1 de novo PTVs partitioned into class 1 de novo PTVs in pLI ≥ 0.9 genes (right), and class 1 de novo PTVs in pLI < 0.9 genes (middle). The entire observed association for de novo PTVs resides in class 1 de novo PTVs in pLI ≥ 0.9 genes. For all such analyses, the rate ratio and significance were calculated by comparing the rate for ID/DD and ASD to the rate in unaffected ASD siblings using a two-sided Poisson exact test28 for synonymous variants and one-sided for the remainder (Online Methods). Error bars represent 95% confidence intervals throughout (a) – (d). See Supplementary Fig. 3 for the corresponding figures using the non-psychiatric version of ExAC. ID/DD, intellectual disability / developmental delay; ASD, autism spectrum disorder; PTV, protein truncating variant; pLI, probability of loss-of-function intolerance; NS, not significant.
