Figure 2.
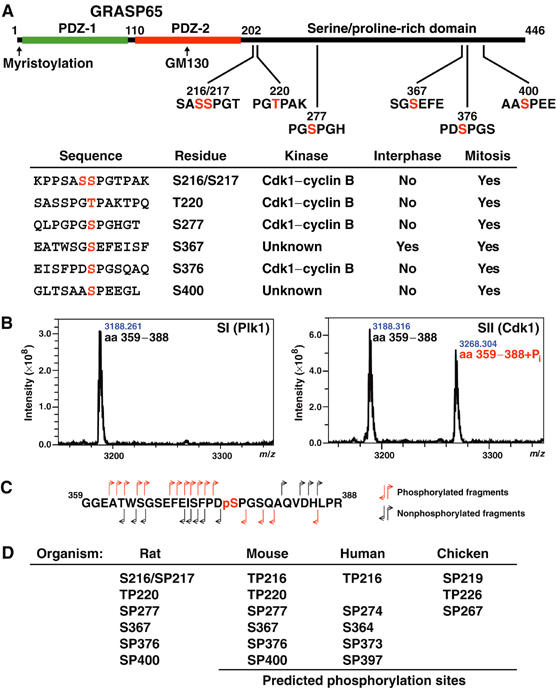
Mapping of phosphorylation sites in GRASP65. (A) Schematic of GRASP65 indicating the two N-terminal PDZ-like domains and GM130 binding region. Phosphorylation sites are shown with the phosphorylated residues numbered and marked in red. The table summarises data on the kinases that phosphorylate these sites and whether or not they are modified in interphase or mitosis. Phosphorylation of S372 was observed under some conditions, but this was substoichiometric and could not be assigned to any kinase with confidence, although it does fall within a Plk1 consensus sequence, and is thus omitted from the table. (B) MALDI-TOF spectra showing a peptide corresponding to GRASP65 amino acids 359–388, phosphorylated with SI and SII from the Superose 6 fractionation of mitotic extract (see Figure 1C). A peak corresponding to the addition of a single phosphate is observed with the Cdk1–cyclin B-containing SII fraction. (C) Fragmentation of the phosphorylated GRASP65359−388 peptide by tandem mass spectrometry gives daughter ions derived from the N- and C-termini of the peptide indicating that serine 376 (pS), shown in red, is the phosphorylated residue. (D) Phosphorylation sites predicted to be equivalent to those identified in rat GRASP65 are shown for mouse, human, and chicken GRASP65.
