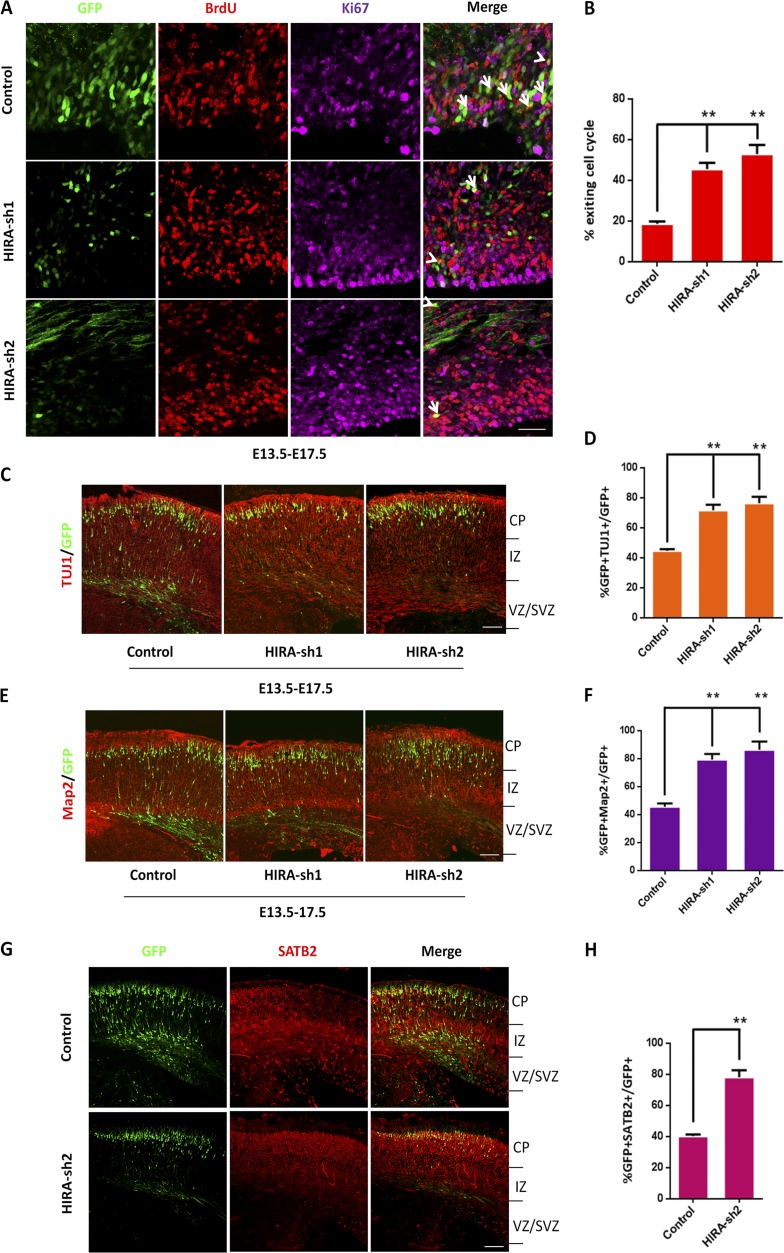
Figure 3.
HIRA loss of function facilitates cell cycle exit and neuronal differentiation. (A and B) HIRA knockdown in NPCs increases premature cell cycle exit in utero. Control or HIRA shRNA plasmids were electroporated into E13.5 mouse brains, and BrdU was injected at E16.5. At E17.5, embryonic brains were collected for immunohistochemical analysis using anti–BrdU and anti–Ki67 antibodies. The cell cycle exit index was calculated by the percentage of GFP-positive cells that exited the cell cycle (GFP+BrdU+Ki67−) divided by the total number of GFP and BrdU double-positive (GFP+BrdU+) cells. The arrows indicate GFP+BrdU+Ki67+ cells, and the arrowheads indicate GFP+BrdU+Ki67− cells. The bar graph shows the percentage of GFP+BrdU+Ki67− cells relative to the total number of BrdU and GFP double-positive cells in the VZ/SVZ (n = 3; mean ± SEM; **, P < 0.01; t test, two sided). Bar, 20 µm. (C and D) HIRA knockdown increases neuronal differentiation in utero. E17.5 brain sections were stained for TUJ1 after the electroporation of control or HIRA-shRNA plasmids into the brain at E13.5. The percentage of GFP and TUJ1 double-positive cells relative to the total number of GFP-positive cells is displayed as a bar graph (n = 3; mean ± SEM; **, P < 0.01; t test, two sided). Bar, 50 µm. (E and F) HIRA knockdown increases neuronal differentiation in utero. E17.5 brain sections were stained for Map2 after the electroporation of control or HIRA-shRNA plasmids into the brain at E13.5. The percentage of GFP and Map2 double-positive cells relative to the total number of GFP-positive cells is displayed as a bar graph (n = 3; mean ± SEM; *, P < 0.05; **, P < 0.01; t test, two sided). Bar, 50 µm. (G and H) HIRA knockdown increases specific neuronal differentiation in utero. E17.5 brain sections were stained for SATB2 after the electroporation of control or HIRA-shRNA plasmids into the brain at E13.5. The percentage of GFP and SATB2 double-positive cells relative to the total number of GFP-positive cells is displayed as a bar graph (n = 3; mean ± SEM; **, P < 0.01; t test, two sided). Bar, 50 µm.