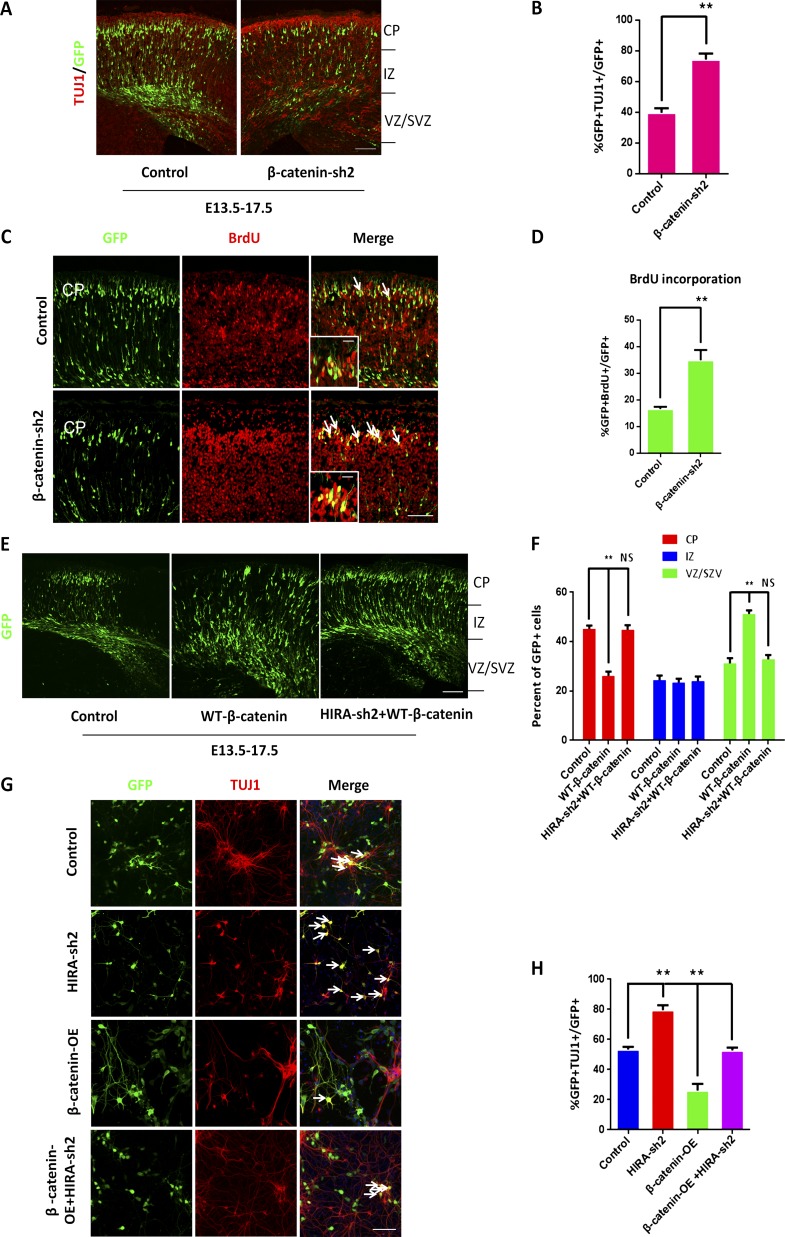
Figure 6.
Overexpression of β-catenin rescues the HIRA knockdown–induced phenotype. (A and B) β-Catenin knockdown increases neuronal differentiation in utero. E17.5 brain sections were stained for anti–TUJ1 antibody after the electroporation of control or β-catenin shRNA plasmids into the brain at E13.5. The percentage of GFP and TUJ1 double-positive cells relative to the total number of GFP-positive cells is displayed as a bar graph (n = 3; mean ± SEM; **, P < 0.01; t test, two sided). Bar, 50 µm. (C and D) β-Catenin knockdown promotes NPC terminal mitosis. Control or β-catenin shRNA plasmids were electroporated into embryonic mouse brains at E13.5, and BrdU was injected at E14.5. Then, the electroporated embryonic brains were collected for immunohistochemical analysis using an anti–BrdU antibody at E18.5. The arrows indicate BrdU and GFP double-positive cells. Insets show high-magnification views. The bar graph shows the percentage of GFP and BrdU double-positive cells relative to the total number of GFP-positive cells in the CP (n = 3; mean ± SEM; **, P < 0.01; t test, two sided). Bars: (main) 25 µm; (insets) 10 µm. (E and F) β-Catenin rescue the positioning defects caused by HIRA knockdown in vivo. Control, WT-β-catenin, or HIRA shRNA together with WT-β-catenin plasmids were electroporated into E13.5 embryonic mouse brains, and embryos were sacrificed at E17.5 for phenotypic analysis. The percentage of GFP-positive cells in each region is exhibited (n = 3; mean ± SEM; **, P < 0.01; NS, not significant; t test, two sided). Bar, 50 µm. (G and H) β-Catenin rescues the proliferative defects caused by HIRA loss of function in vitro. NPCs were isolated from E12.5 embryonic mouse brains and cultured for 1 d. Subsequently, NPCs were infected with control, HIRA shRNA, β-catenin overexpression, or HIRA shRNA together with β-catenin overexpression lentivirus.The arrows show some GFP-positive cells. The percentage of TUJ1and GFP double-positive cells divided by the total number of GFP-positive cells is shown as a bar graph (n = 3; mean ± SEM; **, P < 0.01; t test, two sided). Bar, 25 µm.