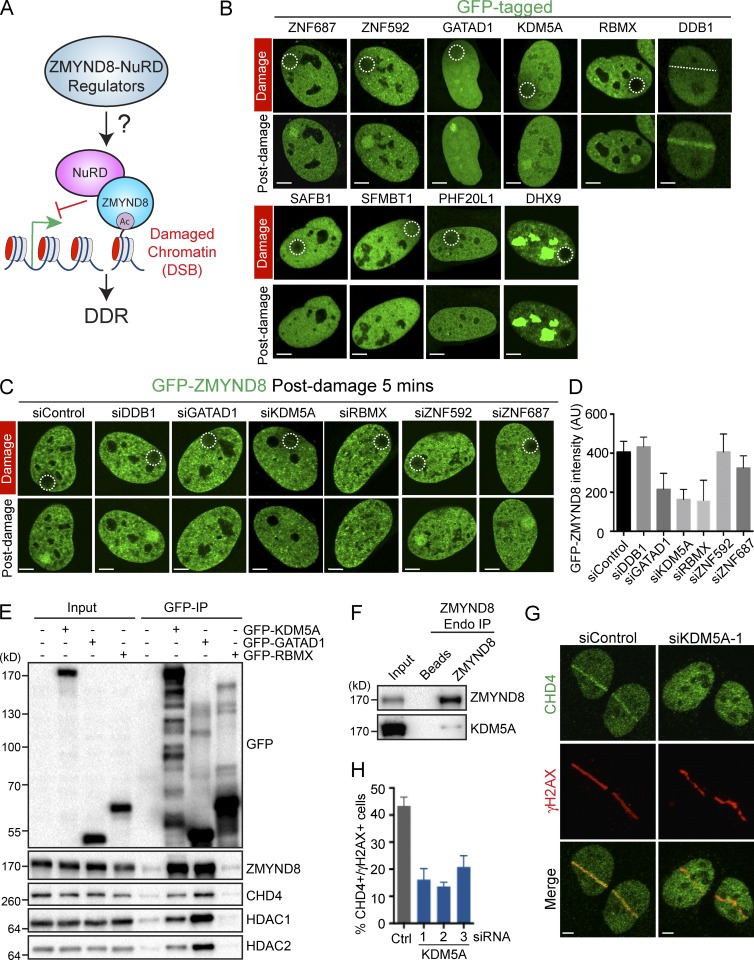
Figure 1.
Identification of ZMYND8–NuRD DDR regulators, including the demethylase KDM5A. (A) Scheme for interrogating the ZMYND8–NuRD DDR pathway. (B) Potential ZMYND8-interacting factors were GFP tagged and screened for damage-dependent relocalization by laser damage with live-cell confocal microscopy. (C) Proteins identified in B were analyzed for their involvement in promoting GFP-ZMYND8 damage localization. U2OS cells stably expressing GFP-ZMYND8 were treated with the indicated siRNAs and analyzed as in B. (D) Quantification of C. Fluorescence intensity (in arbitrary units [AU]) of GFP-ZMYND8 in damaged versus undamaged regions at 5 min after damage is plotted. One representative experiment out of two is shown (error bars indicate SEM, n > 10 cells per condition). (E) GFP-tagged proteins were expressed in HEK293T cells and purified with GFP-TRAP beads. Samples were analyzed by WB with the indicated antibodies. (F) Analysis of endogenous ZMYND8 and KDM5A interactions. ZMYND8 was immunoprecipitated from 293T cells, and samples were analyzed as in E. (G) Recruitment of CHD4 to laser damage in siControl and siKDM5A U2OS cells. (H) Quantification of G. Data represent analysis of >50 cells; n = 2. Error bars represent SEM. Regions damaged are indicated by dotted white lines/circles. Bars, 5 µm.