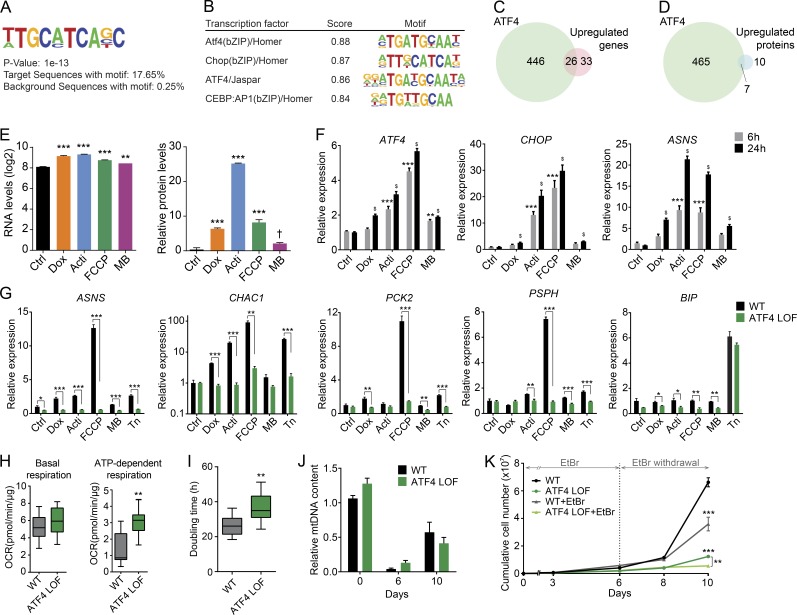
Figure 6.
ATF4 mediates the mitochondrial stress response. (A) Consensus de novo motif identified using HOMER software by selecting the common up-regulated genes in all stress conditions. (B) Representation of the common transcription factors that match with the identified motif. (C and D) Venn diagram showing the similarity between the ATF4 targets and the common induced (C) transcripts and (D) proteins reflecting mitochondrial stress in RNA sequencing and proteomics analysis, respectively. ATF4 chromatin immunoprecipitation and sequence: GSE35681. (E) Relative mRNA, represented as normalized counts, and relative protein levels of ATF4. Data were extracted from RNA sequencing and proteomics, respectively. †, P < 0.1; **, P < 0.01; ***, P < 0.001. (F) Transcript levels of ATF4, CHOP (DDIT3), and ASNS upon 6 and 24 h of exposure to the different mitochondrial stressors. Expression of ATF4 and its targets genes showed time-dependence activation. **, P < 0.01; ***, P < 0.001 relative to control at 6 h; $, P < 0.001 relative to control at 24 h. (G) mRNA expression analysis of ATF4 targets, ASNS, CHAC1, PCK2, PSPH, and the ER stress marker BIP upon 6 h of treatment with the different mitochondrial stressors and the ER stressor tunicamycin (Tn at 2.5 µg/ml) in WT and ATF4 LOF HeLa cells. (H) Boxplots showing the basal and ATP-dependent respiration of ATF4 LOF HeLa cells. OCR, oxygen consumption rate. (I) Doubling time in hours of WT and ATF4 LOF HeLa cells. (J) Relative mtDNA levels of WT and ATF4 LOF HeLa cells after 6 d of 50 ng/ml EtBr treatment and 4 d of recovery (10 d). (K) Cumulative cell number of WT and ATF4 LOF HeLa cells treated or untreated with EtBr for 10 d (6 d with EtBr and 4 d of recovery). All experiments were independently performed at least two times, using triplicates for each condition; data are presented as mean ± SEM of a representative experiment, *, P < 0.05; **, P < 0.01; ***, P < 0.001. Acti, actinonin; Dox, doxycycline; MB, MitoBloCK-6.