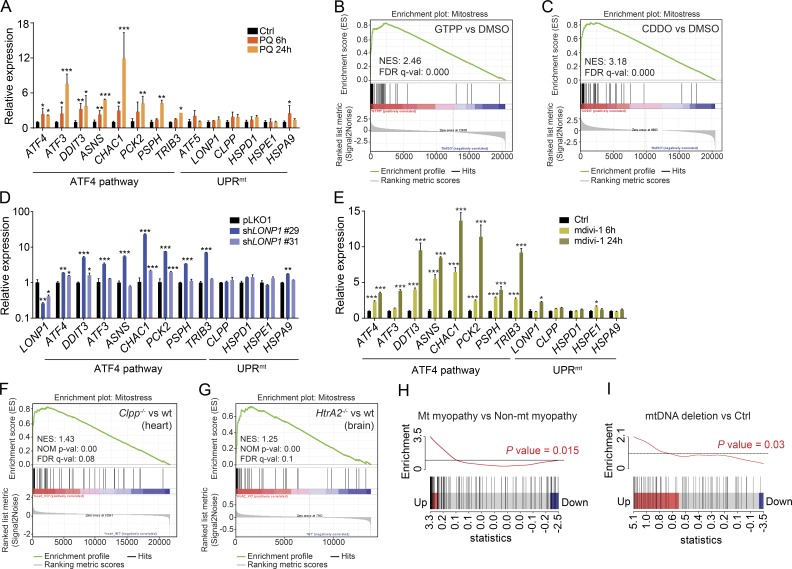
Figure 8.
Mitochondrial stress activates the ATF4 pathway in vitro and in vivo independent of the UPRmt. (A) mRNA expression analysis of the ATF4 pathway and UPRmt genes in HeLa cells 6 and 24 h after exposure to paraquat (PQ) at a final concentration of 400 µM. (B and C) Enrichment score plots from GSEA using RNA sequencing data of cells treated with (B) the TRAP1 inhibitor GTPP and (C) the LONP1 inhibitor CDDO (GSE75247). List of mitochondrial stress genes (mt-stress genes; Table S5) was used as the gene set of interest. FDR q-val, false discovery rate adjusted p-value; NES, normalized enrichment score. (D and E) mRNA expression analysis of the ATF4 pathway and UPRmt genes in HeLa cells after (D) knockdown of LONP1 using two different shRNAs and (E) 6 and 24 h of exposure to mdivi-1 at a final concentration of 50 µM. (F and G) Enrichment score plots from GSEA using microarray data (F) from hearts of mice deficient in ClpP (GSE40207) and (G) from brains of mice deficient in Htra2 (GSE13035). NOM p-val, nominal p-value. (H and I) Barcode plot representing the enrichment in the core mt-stress genes in (H) mitochondrial myopathies versus nonmitochondrial myopathies (GSE43698) and (I) myopathies caused by mtDNA deletions (GSE1462). Enrichment was analyzed using the fold-change relative to control. p-value was calculated using the ROAST test, which confirmed that gene expression changes induced by mitochondrial stress significantly correlate with gene expression changes caused by mitochondrial myopathies. Black vertical lines represent the genes, and the red and blue rectangles the cutoff for the up-regulated and down-regulated genes, respectively. The black horizontal line indicates neutral enrichment, whereas the red line shows the enrichment of up-regulated genes. All experiments were independently performed at least two times, using triplicates for each condition; data are presented as mean ± SEM of a representative experiment; *, P < 0.05; **, P < 0.01; ***, P < 0.001.