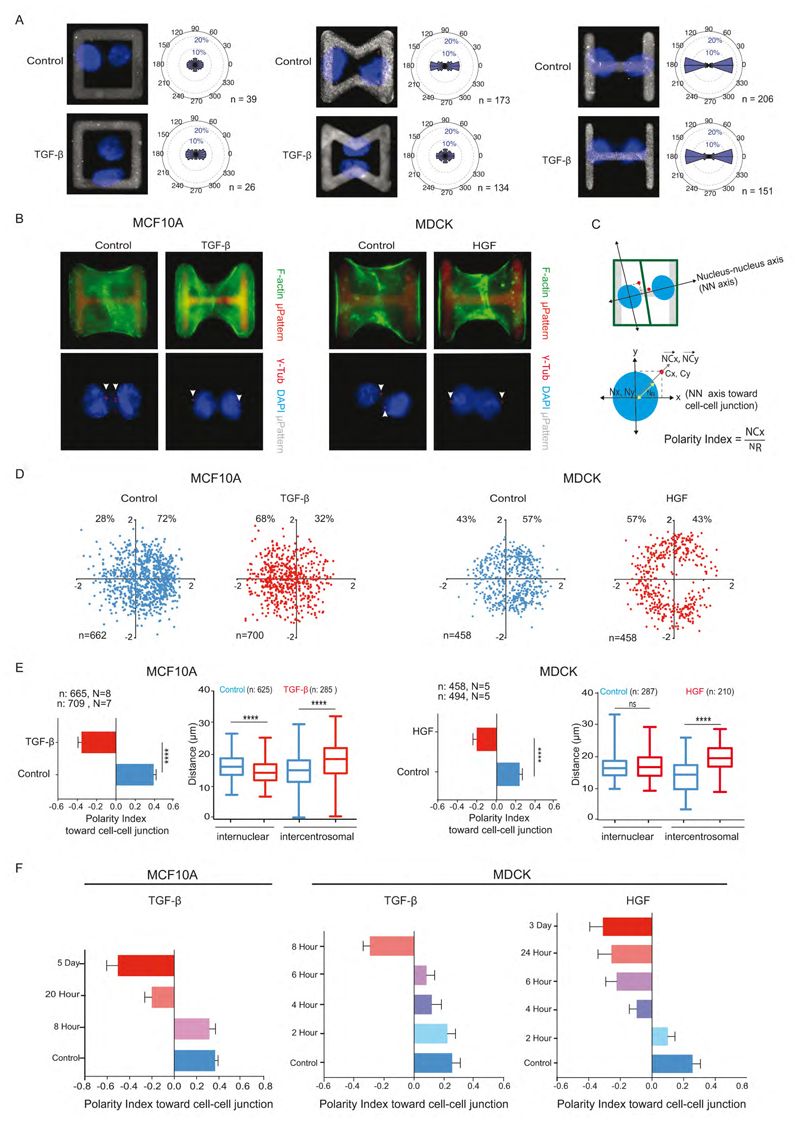
Figure 2. Polarity reversal is an early feature of EMT.
(A) Images of nuclei (blue) of MCF10A cell doublets on square (i), bowtie (ii) and H-shaped (iii) microcropatterns (grey). Graphs represent angular distribution of nucleus-nucleus axis (NN axis) orientation of cell doublets. n indicates number of cells.
(B) MCF10A and MDCK cell doublets on H-shaped micropattern were stained for F-actin (green) (top) or centrosome (red) and DNA (blue) (bottom).
(C) Axes system defined by NN axis (X axis) passing through center of nuclei of cell doublets and an axis perpendicular to NN axis (Y axis). Normalized nucleus-centrosome vector coordinates (NCx, NCy) were calculated by subtracting coordinates of centrosome (Cx, Cy) from Nucleus (Nx, Ny) and normalized by the nucleus distance (NR).
(D) Scatter plot of normalized NC vector. The total number of cells and the respective proportions (%) on positive and negative x-axis are indicated.
(E) Horizontal histograms show the quantification of polarity index, i.e normalized X coordinate of NC vector. N indicates the number of independent experiments, whereas n indicates the total number of single cells. Vertical box plots show the quantification of inter-nuclear and inter-centrosome distance.
(F) Polarity index toward cell-cell junction in control (blue) after varying the duration of TGF-β treatment and HGF treatment to MCF10A and MDCK cells.
Two tailed-non-parametric Mann-Whitney tests were used. ****: p<0.0001. Error bars indicate SEM. See also Figure S2 and S3.