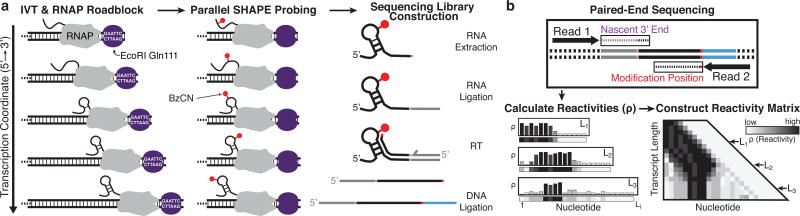
Fig. 1.
Cotranscriptional SHAPE-Seq overview. (a) A set of templates are generated that each contain an E. coli promoter, a variable length RNA template, and an EcoRI Gln111 roadblock site. Single-round in vitro transcription (IVT) with E. coli RNA polymerase (RNAP) is performed using a template library containing a roadblock site at each intermediate transcript length, followed by simultaneous SHAPE probing of the arrested complexes and preparation for sequencing. (b) Paired-end sequencing reveals the SHAPE modification position and the 3’ end of each nascent RNA transcript. Reads are binned by transcript length and used to calculate SHAPE reactivity profiles that are stacked to generate the reactivity matrix. Increases or decreases in reactivity between transcript lengths (rows) at particular nts (columns) of this matrix reveal cotranscriptional folding events.