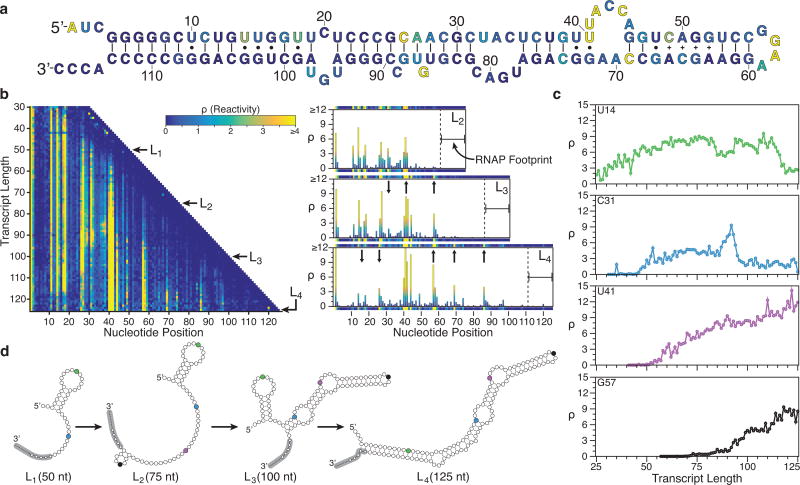
Fig. 2.
SRP RNA cotranscriptional folding. (a) Secondary structure of the final SRP RNA fold colored by reactivity intensity at length L4 (125 nts) drawn according to a crystal structure determined in Batey et al.23. (b) Cotranscriptional SHAPE-Seq reactivity matrix for the SRP RNA folding pathway (left). Lengths L1, L2, L3, and L4 correspond to 50, 75, 100, and 125 nts, respectively. Selected bar charts and corresponding matrix rows above and below (right) display reactivities for L2 to L4. Reactivity changes for L2 → L3 and for L3 → L4 are marked with arrows. (c) Reactivity values for positions U14, C31, U41, and G57 over the course of transcription. U14 undergoes a loop → helix transition at length 117. Similarly, C31 becomes paired at length 96. Plot colors correspond to the marked base positions in (d) outlining a proposed folding pathway of the SRP RNA that is consistent with these transitions. The 14 nt RNAP footprint24 for each length is indicated with gray, with the ~5 nts in the RNA exit channel marked as small circles. Results shown in b and c are n=1 and are representative of three biological replicates (Supplementary Fig. 9a).