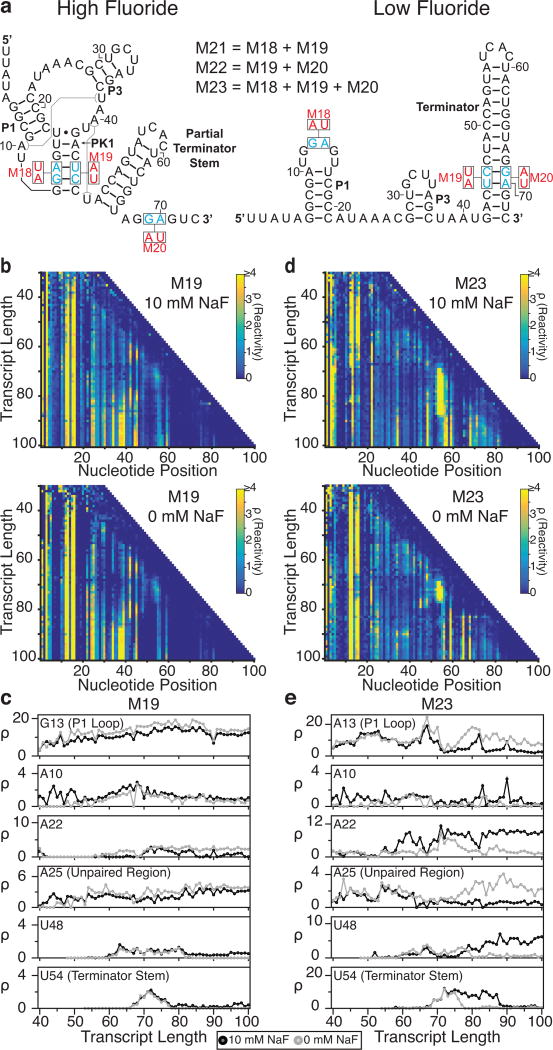
Fig. 5.
Cotranscriptional SHAPE-Seq analysis of B. cereus fluoride riboswitch mutants. (a) The locations of mutations M18-M23 from Baker et al.26 within the antiterminated (high fluoride) and terminated (low fluoride) secondary structures for the wild-type system (see Fig. 3a). (b) Reactivity matrices of the M19 mutant (pseudoknot and terminator base pairing disrupted) transcribed with either 10 mM (top) or 0 mM NaF (bottom). (c) Single nucleotide cotranscriptional reactivity trajectories for the same key nucleotides highlighted in Fig. 4 for the M19 mutant when transcribed with either 0 mM (gray) or 10 mM NaF (black). (d) As in (b), but for mutant M23 (pseudoknot and terminator base pairing restored through compensatory mutations). (e) As in (c), but for mutant M23. Results shown are n=1.