Extended Data Figure 1.
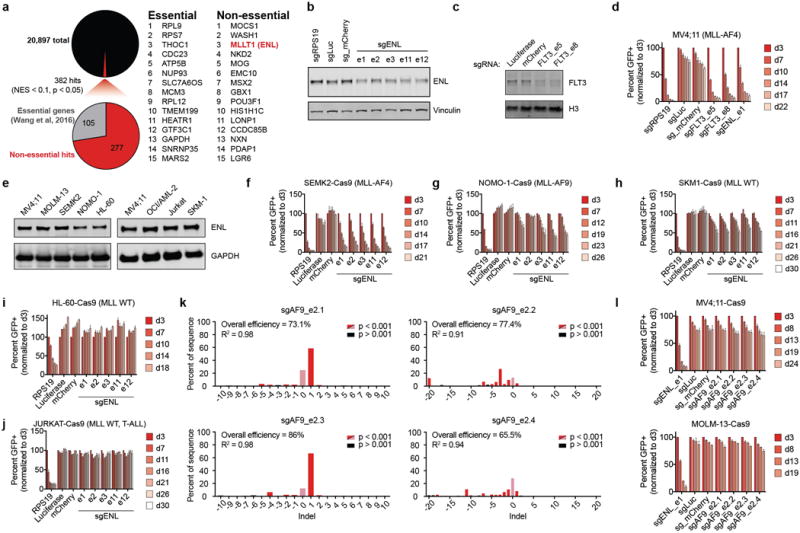
(a) Summary of GeCKOv2 screen results in MV4;11 cells showing the number of hits (NES < 0.1 and p < 0.05 in both replicates) and the percentage of hits known to be essential genes. Right: Top fifteen hits in essential and non-essential protein-coding genes.
(b) Immunoblot of ENL 5 days after transduction with the indicated sgRNA.
(c) Immunoblot of FLT3 5 days after transduction with the indicated sgRNA.
(d) Competition-based CRISPR-Cas9 mutagenesis in MV4;11-Cas9 cells. Percent GFP+ subpopulation (sgRNA-positive) after transduction with lentiviral constructs co-expressing GFP and an sgRNA indicated. Mean ± s.d., n = 3.
(e) ENL protein expression by immunoblot of all cell lines tested for ENL-dependent growth.
(f–j) Same as in (d) for the cell lines and sgRNAs indicated.
(k) Indel quantification by TIDE (tracking of indels by sequence trace decomposition) analysis 5 days after transduction with the sgRNA indicated in MV4;11-Cas9 cells.
(l) Same as in (d) for the cell lines and sgRNAs indicated, n = 4 for MV4;11, n = 3 for MOLM-13.
