Figure 1. ENL is required for growth of acute leukemia.
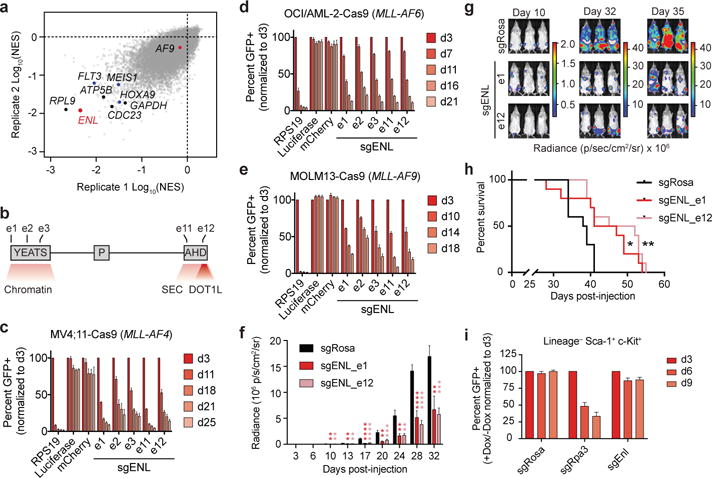
(a) RIGER analysis (normalized enrichment score, NES) of pooled CRISPR/Cas9 screen in MV4;11 cells. Select essential and leukemic driver genes are shown in black and blue, respectively.
(b) Schematic depiction of ENL domain structure: YEATS, polyproline region (P), and AHD (Anc1 homology domain). Numbering indicates sgRNA exon targets in (c-e).
(c–e) Competition-based CRISPR-Cas9 mutagenesis assays. GFP+ (sgRNA+) subpopulation percentage is depicted for the indicated day after lentiviral transduction. Mean ± s.d., n = 3.
(f) Luminescence of MV4;11 (Cas9+, Luciferase+) cells in a disseminated model of leukemia at the indicated day after xenotransplantation. sgRosa denotes a control sgRNA targeting the mouse Rosa26 locus (non-targeting in human cells). P values obtained via two-tailed Student’s t-tests, comparing each ENL sgRNA against the Rosa26 sgRNA. Mean ± s.e.m., n = 10.
(g) Representative images of mice from (f). The same mice are depicted at each time-point.
(h) Kaplan–Meier plot for overall survival of mice from (f). n = 10, p-value obtained from Mantel-Cox test.
(i) Competition-based CRISPR-Cas9 mutagenesis in LSK cells. Percent GFP+ (sgRNA+) after doxycycline induction of Cas9 expression. Mean ± s.d., n = 3.
**** p-value < 0.0001, *** p-value < 0.001, ** p-value < 0.01, * p-value < 0.05.
