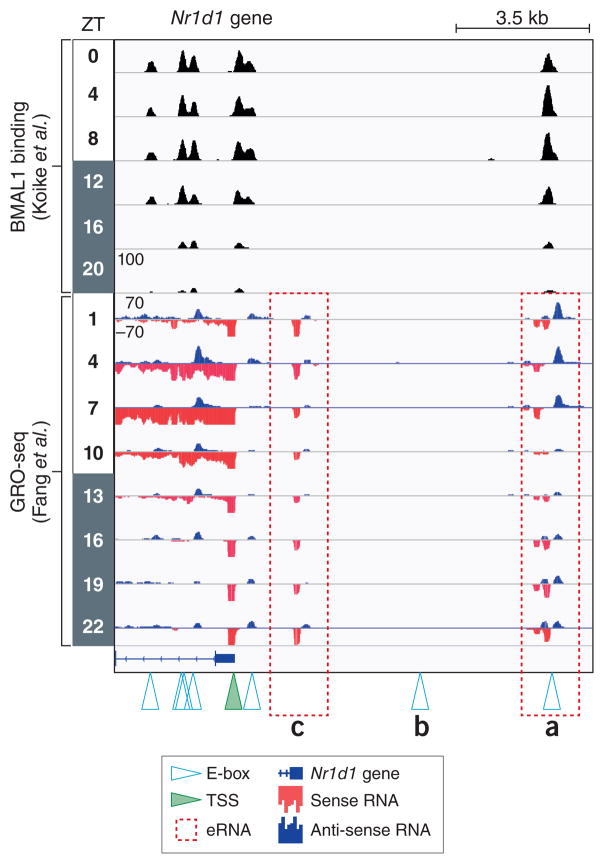
Figure 1. The Nr1d1 gene locus.
ChIP-seq data from Koike et al.8 juxtaposed to Global Run-On Sequencing (GRO-Seq) data from Fang et al.11 performed in mouse livers from circadian time points (ZT0 is light on, ZT 12 is lights off). Canonical CACGTG E-box motifs (denoted by open triangles) are bound by BMAL1 with the exception of (b). GRO-seq measures nascent transcription levels of genic and intergenic RNA, including short-lived enhancer RNA (eRNA). Two rhythmically expressed eRNAs (a) and (c) are present near the transcriptional start site (TSS, green triangle) of Nr1d1, but only one (a) is directly bound by BMAL1. Whether these eRNA are linked topologically or whether the eRNA in (c) is independent of BMAL1 activity is not known.