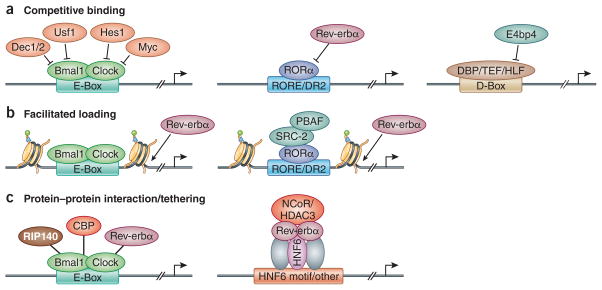
Figure 2. Mechanisms regulating genomic binding of circadian TFs.
(a) Competitive binding among different TFs is observed at E-box, RORE, and D-box motifs. TF binding to the E-box motif, including DEC1/2, USF1, HES1, MYC can inhibit BMAL1 binding. Circadian expressed Rev-erbα competes with RORα, which results in oscillation of RORα binding. E4BP4 competes with other PAR-domain basic leucine zipper TFs DBP/TEF/HLF, which also leads to a robust diurnal switch on/off of their target gene transcription. (b) BMAL1 and CLOCK can function as pioneer TFs to promote the chromatin de-condensation during activation phase. Similarly, RORα could also recruit epigenetic modification machinery to remodel the chromatin structure to facilitate the binding of Rev-erbα. (c) PXDLS peptide motif, which is conserved in CBP, RIP140 and Rev-erbα, can interact with BMAL1/CLOCK and regulate circadian transcription. On the other hand, Rev-erbα can also be tethered to lineage determining TFs, such as HNF6 in the liver, to regulate different metabolic output processes in various tissues.