Fig. 2.
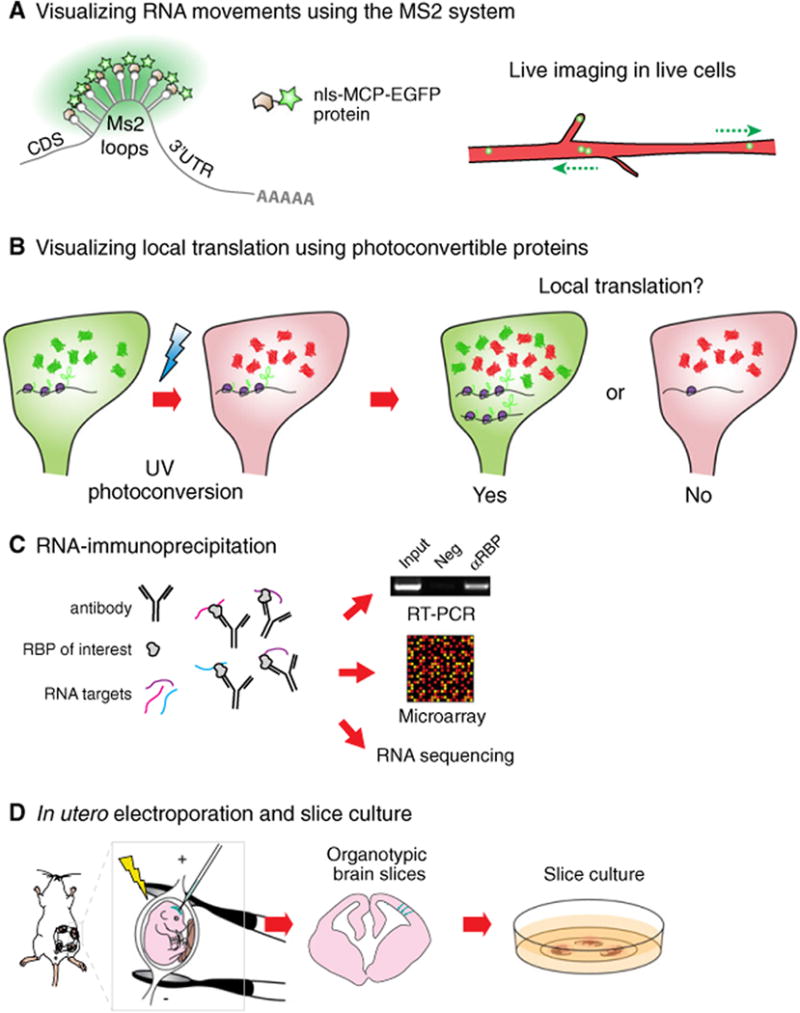
Tools used to study RNA localization and local translation in the developing mouse cortex. (A) In the Ms2 system, DNA constructs expressing an RNA bearing Ms2 loops and a localization sequence are introduced into cells together with Ms2-loop coat protein fused to EGFP (MCP-EGFP). A nuclear localization signal (nls) is added to the MCP-EGFP to reduce background signal of unbound protein in the cytoplasm, and ensure that local events outside the nucleus are actively controlled. RNAs can harbor 24 EGFP molecules, which is sufficient to detect single transcripts. (B) Photoconvertible proteins such as DENDRA2 or KAEDE are valuable to visualize local translation in a distal region of a cell. Reagents expressing RNAs bearing the protein expression cassette together with a localization signal are introduced into cells. After photoconversion, local recovery of the green signal over time indicates local translation (YES), whereas no new green signal suggests the absence of local translation (NO). (C) RNA-immunoprecipitation allows for the identification of RNA targets of RBPs. RNAs pulled down using an antibody targeting a candidate RBP can be analyzed using RT-PCR, microarray or RNA sequencing analyses. This approach has many modifications including crosslinking. (D) In utero electroporation allows for the transfection of RGCs in vivo. Briefly, nucleic acids or proteins are injected into the lateral ventricle of embryonic brains. An electric current is applied to make cell membranes permeable and to direct nucleic acids or proteins into the cells lining the ventricular border. Following electroporation, organotypic brain slices can be cultured for up to several days and subsequently used for live imaging of electroporated cells.
