Fig. 1. Multiply-nicked circles do not enter pulsed-field gel.
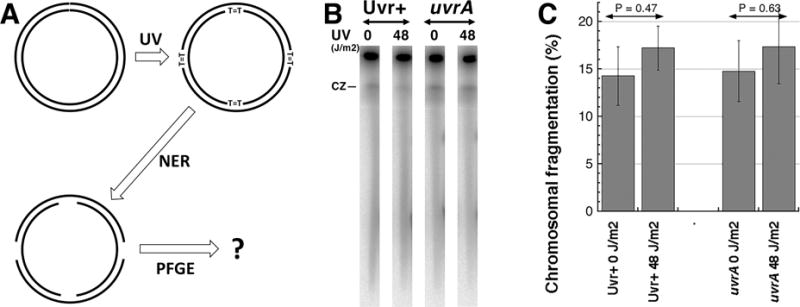
A. A scheme of the experiment: early-stationary cells (so that most chromosomes have no replication bubbles), grown with 32P orthophosphoric acid to label chromosomal DNA, are irradiated with high UV doses (introducing ~1,500 pyrimidine dimers per genome equivalent) as described [7,11]. Following UV exposures, the cells were diluted in spent LB to keep replication inhibited and, after giving 10 minutes at 37°C for nucleotide excision repair (NER) to initiate lesion removal, their chromosomal DNA is isolated in agarose plugs and subjected to pulsed-gel electrophoresis to reveal chromosomal fragmentation [7,8].
B. A representative pulsed-field gel of NER+ (AB1157) versus NER− (uvrA mutant, SRK303-1) cells. CZ, compression zone.
C. Quantification of several gels like in “B”. Chromosomal fragmentation is quantified as the percentage of the total DNA (well + lane) found in the lane. The data are averages ± SEM, with n = 4 or 5. P is a two-tail probability that two values are different (should be 0.05 or less for them to be considered different).
