Fig. 1. Duplication and calibration nodes in the phylogenetic gene tree topologies.
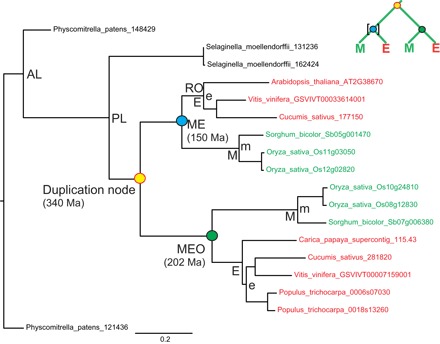
Example of a gene tree with (ME)(ME) topology, tree RAxML_1111 from Jiao et al. (5), in which both paralogs were retained in both monocots (M) and eudicots (E) after the duplication event. Age estimates of nodes were extracted from the original r8s output file of Jiao et al. (5) and are given in parentheses for colored nodes. The nodes for the split of bryophytes (AL node), lycophytes (PL node), monocots and eudicots (ME and MEO nodes), and rosids (RO node) were also extracted from the original r8s output file. The green MEO node was the uncalibrated ME node in the r8s analysis of Jiao et al. (5) (indicated by the absence of square brackets in the small schematic tree at the top right). M and E nodes represent the crown nodes of monocots and eudicots, respectively. m and e nodes are additional calibration nodes that were used when testing the potentially too young upper constraint for the ME nodes (see Methods for details). Examples of gene trees with (ME)(M) and (ME)(E) topologies can be found in fig. S1
