Figure 2.
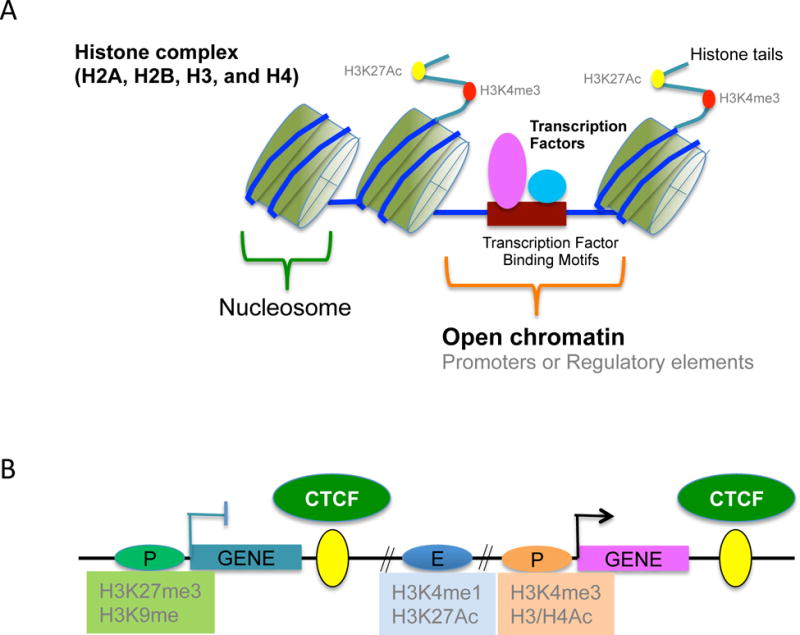
A. The core proteins of nucleosomes are two copies of H2A, H2B, H3, and H4 and 146 base pairs of DNA wraps around these core histone octamer. Core histones are highly conserved and have amino-terminal tails which are subject to various post-translational modifications. Histone modifications, such as methylation and acetylation, play an important role in gene expression and active promoter regions are distinguished by specific histone modifications including H3K4me3 and H3K27Ac. Nucleosomes are depleted in highly active regions, called “open chromatin”. Transcription factors and lineage determining factors such as NFAT and RUNX2 bind to a specific binding motif in promoters or enhancers. Ac, acetylation; Me, methylation.
B. Histone modifications of functional elements including promoters, enhancers, and insulators. Insulator is a genetic boundary element that blocks the interaction between enhancers and promoters and is defined by CCCTC-binding factor (CTCF) binding although CTCF has dual effects on enhancers; either blocking or activating. Active promoters are enriched for H3K4me3 and H3/H4 acetylation. H3K9 me2/3 and H3K27me3 are associated with repressed promoter regions. Active enhancers are marked by H3Kme1 and H3K27Ac.
